

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146704.8 - phase: 1 /pseudo
(946 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
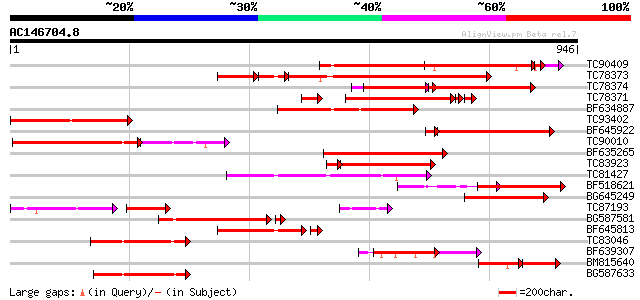
Score E
Sequences producing significant alignments: (bits) Value
TC90409 weakly similar to GP|10280097|emb|CAC09980. nucleotide s... 497 e-142
TC78373 weakly similar to GP|14348622|gb|AAK61318.1 NBS-LRR resi... 402 e-140
TC78374 236 7e-86
TC78371 similar to GP|9558523|dbj|BAB03441.1 ESTs AU078742(C1188... 267 3e-79
BF634887 weakly similar to GP|10280097|em nucleotide sequence of... 270 1e-72
TC93402 weakly similar to GP|16588610|gb|AAL26857.1 NBS-type put... 268 1e-71
BF645922 239 1e-69
TC90010 similar to GP|5817349|gb|AAD52718.1| putative NBS-LRR ty... 183 3e-64
BF635265 similar to GP|4234955|gb|A NBS-LRR-like protein cD8 {Ph... 238 8e-63
TC83923 weakly similar to GP|14348622|gb|AAK61318.1 NBS-LRR resi... 214 7e-61
TC81427 weakly similar to GP|11994216|dbj|BAB01338. disease resi... 226 2e-59
BF518621 weakly similar to GP|11994216|db disease resistance com... 206 3e-53
BG645249 191 9e-49
TC87193 similar to GP|21616922|gb|AAM66423.1 NBS-LRR protein {Or... 134 5e-47
BG587581 weakly similar to GP|11994216|db disease resistance com... 178 1e-44
BF645813 weakly similar to GP|14348622|gb NBS-LRR resistance-lik... 163 3e-42
TC83046 weakly similar to GP|10280097|emb|CAC09980. nucleotide s... 166 3e-41
BF639307 similar to GP|11994216|db disease resistance comples pr... 159 6e-39
BM815640 weakly similar to GP|14348619|gb NBS-LRR resistance-lik... 103 4e-38
BG587633 similar to GP|20385442|gb| resistance gene analog {Viti... 154 2e-37
>TC90409 weakly similar to GP|10280097|emb|CAC09980. nucleotide sequence of
the I-2 resistance gene {Fusarium oxysporum}, partial
(5%)
Length = 1136
Score = 497 bits (1279), Expect(2) = e-142
Identities = 251/364 (68%), Positives = 291/364 (78%), Gaps = 3/364 (0%)
Frame = +3
Query: 518 HMKFDGGREEMDESMAESNVSVLEALQPNRNLKRLTISKYKGNSFPNWIRGYHLPNLVSL 577
+MK+ G +++ + +ESNV EALQPN NL RL IS+YKG SFP WIRG HLPNLVSL
Sbjct: 3 NMKY-GDNYKLNNNRSESNV--FEALQPNNNLNRLYISQYKGKSFPKWIRGCHLPNLVSL 173
Query: 578 NLQFCGLCSLLPPLGTLPFLKMLSISDCDGIKIIGEEFYDSSSINVLFRSLEVLKFEKMN 637
LQ CG C LPPLG LP LK L+I DC GIKIIGEEF+ ++S NV F SLEVLKF KMN
Sbjct: 174 KLQSCGSCLHLPPLGQLPCLKELAICDCHGIKIIGEEFHGNNSTNVPFLSLEVLKFVKMN 353
Query: 638 NWEEWLCLEGFPLLKELYIRECPKLKMSLPQHLPSLQKLFINDCKMLEASIPNGDNIIDL 697
+WEEWLCLEGFPLLKEL I+ CP+L+ +LPQHLPSLQKL I DC++LEASIP GDNII+L
Sbjct: 354 SWEEWLCLEGFPLLKELSIKSCPELRSALPQHLPSLQKLEIIDCELLEASIPKGDNIIEL 533
Query: 698 DIKRCDRILVNELPTSLKKLFILENRYTEFSVEQIFVNSTILEVLELDLNGSLKCPTLDL 757
D++RCD IL+NELPTSLK+ EN + +FSVEQI +N+TILE L+ D GS+KC +LDL
Sbjct: 534 DLQRCDHILINELPTSLKRFVFRENWFAKFSVEQILINNTILEELKFDFIGSVKCLSLDL 713
Query: 758 CCYNSLGELSITRWCSSSLSFSLHLFTNLYSLWFVDCPNLDSFPEGGLPCNLLSLTITNC 817
CY+SL +LSIT W SSSL LHLFTNL+SL +CP LDSFP GGLP NL L I NC
Sbjct: 714 RCYSSLRDLSITGWHSSSLPLELHLFTNLHSLKLYNCPRLDSFPNGGLPSNLRGLVIWNC 893
Query: 818 PKLIASRQEWG---LKSLKYFFVCDDFENVESFPKESLLPPTLSYLNLNNCSKLRIMNNE 874
P+LIA RQEWG L SLK FFV D+FENVESFP+ESLLPPTL+YLNLNNCSKLRIMNN+
Sbjct: 894 PELIALRQEWGLFRLNSLKSFFVSDEFENVESFPEESLLPPTLTYLNLNNCSKLRIMNNK 1073
Query: 875 GFLH 878
GFLH
Sbjct: 1074GFLH 1085
Score = 44.7 bits (104), Expect = 2e-04
Identities = 67/261 (25%), Positives = 112/261 (42%), Gaps = 29/261 (11%)
Frame = +3
Query: 693 NIIDLDIKRCDRIL----VNELPTSLKKLFILENRYTEFSVEQIFVN-STILEVLELDLN 747
N++ L ++ C L + +LP LK+L I + + E+ N ST + L L++
Sbjct: 159 NLVSLKLQSCGSCLHLPPLGQLPC-LKELAICDCHGIKIIGEEFHGNNSTNVPFLSLEVL 335
Query: 748 GSLKCPTLD--LCC--YNSLGELSITRWCSSSLSFSLHLFTNLYSLWFVDCPNLD-SFPE 802
+K + + LC + L ELSI + HL +L L +DC L+ S P+
Sbjct: 336 KFVKMNSWEEWLCLEGFPLLKELSIKSCPELRSALPQHL-PSLQKLEIIDCELLEASIPK 512
Query: 803 GGLPCNLLSLTITNCPKLIASRQEWGLKSLKY---FFVCDDFENV-------ESFPKESL 852
G N++ L + C ++ + LK + +F E + E + +
Sbjct: 513 GD---NIIELDLQRCDHILINELPTSLKRFVFRENWFAKFSVEQILINNTILEELKFDFI 683
Query: 853 LPPTLSYLNLNNCSKLRIMNNEGF--------LHL-KSLEFLYIINCPSLERLPEEALPN 903
L+L S LR ++ G+ LHL +L L + NCP L+ P LP+
Sbjct: 684 GSVKCLSLDLRCYSSLRDLSITGWHSSSLPLELHLFTNLHSLKLYNCPRLDSFPNGGLPS 863
Query: 904 SLYSLWIKDCPLIKVKYQKEG 924
+L L I +CP + Q+ G
Sbjct: 864 NLRGLVIWNCPELIALRQEWG 926
Score = 29.6 bits (65), Expect(2) = e-142
Identities = 13/19 (68%), Positives = 16/19 (83%)
Frame = +1
Query: 876 FLHLKSLEFLYIINCPSLE 894
F LKSL+ LYI++CPSLE
Sbjct: 1078 FSTLKSLKDLYIVDCPSLE 1134
>TC78373 weakly similar to GP|14348622|gb|AAK61318.1 NBS-LRR resistance-like
protein J78 {Phaseolus vulgaris}, partial (7%)
Length = 1463
Score = 402 bits (1033), Expect(3) = e-140
Identities = 208/343 (60%), Positives = 247/343 (71%), Gaps = 4/343 (1%)
Frame = +1
Query: 466 FIVEEQNVSDLKELAKLNHLHGAIDIEGLGNVSDLADSATVNLKDTKYLEEL----HMKF 521
F+V EQ+ SD+KEL LNHL G + I G+ +V D D+A NLKD K++EEL KF
Sbjct: 460 FVVGEQSGSDIKELGNLNHLQGELCISGMEHVIDPVDAAGANLKDKKHVEELIWSGSYKF 639
Query: 522 DGGREEMDESMAESNVSVLEALQPNRNLKRLTISKYKGNSFPNWIRGYHLPNLVSLNLQF 581
+ E D V EALQPN +LKRL IS YKGN F NW+RG LPNLVS+ L
Sbjct: 640 NTNGRESD---------VFEALQPNSSLKRLIISHYKGNRFANWMRGCDLPNLVSIRLNL 792
Query: 582 CGLCSLLPPLGTLPFLKMLSISDCDGIKIIGEEFYDSSSINVLFRSLEVLKFEKMNNWEE 641
C LCS LPPLG LP LK +SIS CD I+IIG+EFY ++S NV FRSLE+L F+ M+ WEE
Sbjct: 793 CALCSELPPLGQLPCLKEISISGCDKIRIIGKEFYGNNSTNVPFRSLEILHFDSMSEWEE 972
Query: 642 WLCLEGFPLLKELYIRECPKLKMSLPQHLPSLQKLFINDCKMLEASIPNGDNIIDLDIKR 701
W LEGFPLLKEL I++CPKLK +LPQHLPSLQKL I DCK LEA IP DN+I+LDIKR
Sbjct: 973 WSHLEGFPLLKELSIKKCPKLKRALPQHLPSLQKLEIKDCKKLEALIPKCDNMIELDIKR 1152
Query: 702 CDRILVNELPTSLKKLFILENRYTEFSVEQIFVNSTILEVLELDLNGSLKCPTLDLCCYN 761
CDRILVNELPT+LK LF+ +N+YTEFSV+Q +N LEVL+ D G + CP+LDL CYN
Sbjct: 1153CDRILVNELPTNLKSLFLCDNQYTEFSVDQNLINILFLEVLKFDFRGCVNCPSLDLRCYN 1332
Query: 762 SLGELSITRWCSSSLSFSLHLFTNLYSLWFVDCPNLDSFPEGG 804
SL +LSI W SSSL LHLFT+L +L CP L+SFP GG
Sbjct: 1333SLRDLSIKGWHSSSLPLELHLFTSLSALRLYHCPELESFPMGG 1461
Score = 84.3 bits (207), Expect(3) = e-140
Identities = 43/69 (62%), Positives = 55/69 (79%)
Frame = +2
Query: 347 LISNNVQQDLFSRLNFLRMLSFRGCGLLELVDEISNLKLLRYLDLSYTWIEILPDTICML 406
+ SNN+Q+DLFS+L +LRMLSF G L EL EI NLKLLRYL+L+ + IE LPD+IC L
Sbjct: 113 MTSNNLQRDLFSKLKYLRMLSFCGFKLKELSGEIGNLKLLRYLNLTASLIERLPDSICKL 292
Query: 407 HNLQTLLLE 415
+ L+TL+LE
Sbjct: 293 YKLETLILE 319
Score = 53.1 bits (126), Expect(3) = e-140
Identities = 29/52 (55%), Positives = 35/52 (66%)
Frame = +3
Query: 415 EGCCELTELPSNFSKLVNLRHLKLPSHNGRPCIKTMPKHTGKLNNLQSLSYF 466
+ C ELT+LPS F K V+LRHL L N IK MPK G+LN+LQ+LS F
Sbjct: 318 KNCFELTKLPSKFYKFVSLRHLHLEGCN----IKKMPKKIGRLNHLQTLSDF 461
Score = 33.9 bits (76), Expect = 0.30
Identities = 56/229 (24%), Positives = 92/229 (39%), Gaps = 4/229 (1%)
Frame = +1
Query: 670 LPSLQKLFINDCKMLEASIPNGDN--IIDLDIKRCDRILVNELPTSLKKLFILENRYTEF 727
LP+L + +N C + P G + ++ I CD+I + I + Y
Sbjct: 760 LPNLVSIRLNLCALCSELPPLGQLPCLKEISISGCDKIRI-----------IGKEFYGNN 906
Query: 728 SVEQIFVNSTILEVLELDLNGSLKCPTLDLCCYNSLGELSITRWCSSSLSFSLHLFTNLY 787
S F + LE+L D + L + L ELSI + + HL +L
Sbjct: 907 STNVPFRS---LEILHFDSMSEWE-EWSHLEGFPLLKELSIKKCPKLKRALPQHL-PSLQ 1071
Query: 788 SLWFVDCPNLDSFPEGGLP-C-NLLSLTITNCPKLIASRQEWGLKSLKYFFVCDDFENVE 845
L DC L++ +P C N++ L I C +++ + LKSL F+CD+ + E
Sbjct: 1072KLEIKDCKKLEAL----IPKCDNMIELDIKRCDRILVNELPTNLKSL---FLCDN-QYTE 1227
Query: 846 SFPKESLLPPTLSYLNLNNCSKLRIMNNEGFLHLKSLEFLYIINCPSLE 894
++L+ N FL + +F +NCPSL+
Sbjct: 1228FSVDQNLI-------------------NILFLEVLKFDFRGCVNCPSLD 1317
>TC78374
Length = 1185
Score = 236 bits (601), Expect(2) = 7e-86
Identities = 121/181 (66%), Positives = 135/181 (73%), Gaps = 3/181 (1%)
Frame = +2
Query: 700 KRCDRILVNELPTSLKKLFILENRYTEFSVEQIFVNSTILEVLELDLNGSLKCPTLDLCC 759
K CDRILVNELPTSLK L + +NRYTEFSV+Q +N LE L+LD G ++CP+LDL C
Sbjct: 602 KSCDRILVNELPTSLKSLLLWQNRYTEFSVDQNLINFPFLESLQLDFTGFVECPSLDLRC 781
Query: 760 YNSLGELSITRWCSSSLSFSLHLFTNLYSLWFVDCPNLDSFPEGGLPCNLLSLTITNCPK 819
YNSL LSI W SSSL LHLFTNL L+ DCP L+SFP GGLP NL SL I NCPK
Sbjct: 782 YNSLWRLSIQGWHSSSLPLELHLFTNLNYLYLYDCPELESFPMGGLPSNLGSLQIYNCPK 961
Query: 820 LIASRQEWG---LKSLKYFFVCDDFENVESFPKESLLPPTLSYLNLNNCSKLRIMNNEGF 876
LI SR+EWG L SLK FFV D+FENVESFP+E+LLPPTL L LN CSKL MNN GF
Sbjct: 962 LIGSREEWGLFQLNSLKSFFVTDEFENVESFPEENLLPPTLETLVLNKCSKLXKMNNXGF 1141
Query: 877 L 877
L
Sbjct: 1142L 1144
Score = 130 bits (327), Expect = 2e-30
Identities = 73/129 (56%), Positives = 87/129 (66%), Gaps = 5/129 (3%)
Frame = +1
Query: 591 LGTLPFLKMLSISDCDGIKIIGEEFYDSSSINVLFRSLEVLKFEKMNNWEEWLCLEGFPL 650
LG LP LK LSI DC+GIKII EEFY ++S V F+SLE L+F M NWEEW+C+ FPL
Sbjct: 7 LGQLPSLKKLSIYDCEGIKIIDEEFYGNNSTIVPFKSLEYLRFVDMVNWEEWICVR-FPL 183
Query: 651 LKELYIRECPKLKMSLPQHLPSLQKLFINDCKMLE--ASIPNGDNIIDLDIKRCD---RI 705
LKELYI CPKLK +LPQHLPSLQKL I CK LE + ++ +L I C R+
Sbjct: 184 LKELYIENCPKLKSTLPQHLPSLQKLIIYGCKELEEWLCLEGFLSLKELSIWYCSKFKRV 363
Query: 706 LVNELPTSL 714
L LP+ L
Sbjct: 364 LPQHLPSLL 390
Score = 101 bits (251), Expect(2) = 7e-86
Identities = 62/132 (46%), Positives = 73/132 (54%)
Frame = +1
Query: 570 HLPNLVSLNLQFCGLCSLLPPLGTLPFLKMLSISDCDGIKIIGEEFYDSSSINVLFRSLE 629
HLP+L L + C L LK LSI C K + + S L
Sbjct: 238 HLPSLQKLIIYGCKELEEWLCLEGFLSLKELSIWYCSKFKRVLPQHLPS---------LL 390
Query: 630 VLKFEKMNNWEEWLCLEGFPLLKELYIRECPKLKMSLPQHLPSLQKLFINDCKMLEASIP 689
L+ N EEWLCL FPLL E I CP+LK +LPQHLPSLQKLFINDC LEASIP
Sbjct: 391 NLRINYCNELEEWLCLGEFPLLNEFSISNCPELKRALPQHLPSLQKLFINDCNKLEASIP 570
Query: 690 NGDNIIDLDIKR 701
DN+I+LDI++
Sbjct: 571 KCDNMIELDIQK 606
Score = 39.3 bits (90), Expect = 0.007
Identities = 19/39 (48%), Positives = 26/39 (65%), Gaps = 1/39 (2%)
Frame = +2
Query: 877 LHL-KSLEFLYIINCPSLERLPEEALPNSLYSLWIKDCP 914
LHL +L +LY+ +CP LE P LP++L SL I +CP
Sbjct: 842 LHLFTNLNYLYLYDCPELESFPMGGLPSNLGSLQIYNCP 958
>TC78371 similar to GP|9558523|dbj|BAB03441.1 ESTs AU078742(C11888)
C26225(C11888) correspond to a region of the predicted
gene.~Similar to Oryza, partial (2%)
Length = 815
Score = 267 bits (683), Expect(4) = 3e-79
Identities = 134/184 (72%), Positives = 151/184 (81%), Gaps = 1/184 (0%)
Frame = +2
Query: 561 SFPNWIRGYHLPNLVSLNLQFCGLCSLLPPLGTLPFLKMLSISDCDGIKIIGEEFYDSSS 620
SFPNWI G HLPNLVSL L CGLCS LPPLG LP LK LSIS CDGIKIIGEEF+ +++
Sbjct: 152 SFPNWITGCHLPNLVSLQLLSCGLCSHLPPLGQLPSLKELSISKCDGIKIIGEEFHGNNN 331
Query: 621 I-NVLFRSLEVLKFEKMNNWEEWLCLEGFPLLKELYIRECPKLKMSLPQHLPSLQKLFIN 679
+ NV F SLEVLKFE MNNWEEWLCLEGFPLLK+L IR CPKLK +LPQHLP LQKL I+
Sbjct: 332 LTNVPFLSLEVLKFEMMNNWEEWLCLEGFPLLKKLSIRNCPKLKRALPQHLPYLQKLNIS 511
Query: 680 DCKMLEASIPNGDNIIDLDIKRCDRILVNELPTSLKKLFILENRYTEFSVEQIFVNSTIL 739
DC +EASIP DN+I+LDI+RCDRILVNELPTSLK LF+ +N YTEFSV+Q +N L
Sbjct: 512 DCNKMEASIPKCDNMIELDIQRCDRILVNELPTSLKWLFLCDNPYTEFSVDQNLINILFL 691
Query: 740 EVLE 743
E L+
Sbjct: 692 EELK 703
Score = 35.0 bits (79), Expect(4) = 3e-79
Identities = 17/35 (48%), Positives = 24/35 (68%)
Frame = +3
Query: 487 GAIDIEGLGNVSDLADSATVNLKDTKYLEELHMKF 521
G + I GL V + D+A NLKD K++EEL+MK+
Sbjct: 6 GKLCISGLEYVINPEDAARANLKDKKHVEELNMKY 110
Score = 32.7 bits (73), Expect(4) = 3e-79
Identities = 16/21 (76%), Positives = 16/21 (76%)
Frame = +1
Query: 759 CYNSLGELSITRWCSSSLSFS 779
CYNSL LSIT W SSSL FS
Sbjct: 751 CYNSLCYLSITGWGSSSLPFS 813
Score = 21.9 bits (45), Expect(4) = 3e-79
Identities = 8/14 (57%), Positives = 10/14 (71%)
Frame = +3
Query: 744 LDLNGSLKCPTLDL 757
LD G + CP+LDL
Sbjct: 705 LDFRGWVNCPSLDL 746
>BF634887 weakly similar to GP|10280097|em nucleotide sequence of the I-2
resistance gene {Fusarium oxysporum}, partial (3%)
Length = 694
Score = 270 bits (691), Expect = 1e-72
Identities = 142/235 (60%), Positives = 176/235 (74%), Gaps = 1/235 (0%)
Frame = +1
Query: 448 KTMPKHTGKLNNLQSLSYFIVEEQNVSDLKELAKLNHLHGAIDIEGLGNVSDLADSATVN 507
K MPK G LN+LQ+LS+F+V E+N S++++L LN L G + I GL +V + D+A N
Sbjct: 1 KKMPKQIGSLNHLQTLSHFVVGEENGSNIQQLGNLNCLQGKLCISGLEHVINPKDAAGAN 180
Query: 508 LKDTKYLEELHMKFDGGREEMDESMAESNVSVLEALQPNRNLKRLTISKYKGNSFPNWIR 567
LKD K++EEL+M++ + + + ESNV EAL PN NLKRL I +Y+GN+FPNWI
Sbjct: 181 LKDKKHVEELNMEYSDNFK-FNNNGRESNV--FEALHPNSNLKRLYIGRYQGNNFPNWIT 351
Query: 568 GYHLPNLVSLNLQFCGLCSLLPPLGTLPFLKMLSISDCDGIKIIGEEFYDSSSINVLFRS 627
G HLPNLVSL L+ CG CS +PP+ LP LK SIS+C+GIKII EEFY +SS NV FRS
Sbjct: 352 GCHLPNLVSLQLKGCG-CSHMPPIEQLPSLKEFSISNCNGIKIITEEFYGNSSTNVSFRS 528
Query: 628 LEVLKFEKMNNWEEWLCLEGFPLLKELYIRECPKLKMS-LPQHLPSLQKLFINDC 681
LEVLKFE+MNNWEEW C EGFPLLKE+YI CPKLK + LPQHLPS+QKL I DC
Sbjct: 529 LEVLKFEEMNNWEEWFCPEGFPLLKEIYIWNCPKLKXALLPQHLPSIQKLKICDC 693
>TC93402 weakly similar to GP|16588610|gb|AAL26857.1 NBS-type putative
resistance protein {Glycine max}, partial (61%)
Length = 676
Score = 268 bits (684), Expect = 1e-71
Identities = 130/204 (63%), Positives = 160/204 (77%)
Frame = +2
Query: 1 YNDNKIKEHFELKAWVYVSESFDVVGLTKAILKSFNSSADGEDLNLLQHQLQYMLMGKKY 60
YND +I+E FELKAWVYVSE FDV+GLT AIL+ F ++ + EDL+LLQ QLQ L GK Y
Sbjct: 68 YNDRRIQETFELKAWVYVSEYFDVIGLTNAILRKFGTAENSEDLDLLQWQLQKKLTGKNY 247
Query: 61 LLVLDDIWNGDAERWELLLLPFNHGSFGSKIVVTTREKEVADNVLKSTELFDLQQLDKSN 120
LLV+DD+W + E WE LLLPFN+GS GSKI++TTR+K+VA ++KSTEL DL+QL +
Sbjct: 248 LLVMDDVWKLNEESWEKLLLPFNYGSSGSKIILTTRDKKVA-LIVKSTELVDLEQLKNKD 424
Query: 121 CWSLFVTHAFQGKSVSEYPNLESVGRKIVEKCGGLPLAIKSLGQLLRKTFSEHEWINILE 180
CWSLF AF G +VSEYP LES+G+ IV+KCGGLPL K++G LLRK F++ EW ILE
Sbjct: 425 CWSLFKRLAFHGSNVSEYPKLESIGKNIVDKCGGLPLTGKTMGNLLRKKFTQSEWEKILE 604
Query: 181 TDMWRLSKVDHNVNSVLRLSYHNL 204
DMWRL+ D N+NS LRLSYHNL
Sbjct: 605 ADMWRLTDDDSNINSALRLSYHNL 676
>BF645922
Length = 658
Score = 239 bits (609), Expect(2) = 1e-69
Identities = 125/198 (63%), Positives = 141/198 (71%), Gaps = 4/198 (2%)
Frame = +3
Query: 715 KKLFILENRYTEFSVEQIFVNSTILEVLELDLNGSLKCPTLDLCCYNSLGELSITRWCSS 774
K + NRYTEFSV Q +N LE LEL+ +GS+KCP+LDL CYN L +LSI WCSS
Sbjct: 63 KSYLLRRNRYTEFSVHQNLINFPFLEALELNWSGSVKCPSLDLRCYNFLRDLSIKGWCSS 242
Query: 775 SLSFSLHLFTNLYSLWFVDCPNLDSFPEGGLPCNLLSLTITNCPKLIASRQEWG---LKS 831
SL LHLFT L SL+ DCP L+S P GGLP NL+ L I NCPKLI SR+EWG L S
Sbjct: 243 SLPLELHLFTKLQSLYLYDCPELESLPMGGLPSNLIQLGIYNCPKLIGSREEWGLFQLNS 422
Query: 832 LKYFFVCDDFENVESFPKESLLPPTLSYLNLNNCSKLRIMNNEGFLHLKSLEFLYIINCP 891
LK F V D+FENVESFP+E+LLPPTL L L NCSKLRIMN + FLHLKSL LYI+ P
Sbjct: 423 LKCFTVADEFENVESFPEENLLPPTLEILQLYNCSKLRIMNKKSFLHLKSLNRLYILX*P 602
Query: 892 SLERLP-EEALPNSLYSL 908
LE LP E LPNSL L
Sbjct: 603 XLESLPXXEDLPNSLSEL 656
Score = 43.9 bits (102), Expect(2) = 1e-69
Identities = 20/23 (86%), Positives = 22/23 (94%)
Frame = +2
Query: 695 IDLDIKRCDRILVNELPTSLKKL 717
I+LDI+ CDRILVNELPTSLKKL
Sbjct: 2 IELDIQNCDRILVNELPTSLKKL 70
Score = 38.5 bits (88), Expect = 0.012
Identities = 19/39 (48%), Positives = 25/39 (63%), Gaps = 1/39 (2%)
Frame = +3
Query: 877 LHL-KSLEFLYIINCPSLERLPEEALPNSLYSLWIKDCP 914
LHL L+ LY+ +CP LE LP LP++L L I +CP
Sbjct: 258 LHLFTKLQSLYLYDCPELESLPMGGLPSNLIQLGIYNCP 374
>TC90010 similar to GP|5817349|gb|AAD52718.1| putative NBS-LRR type disease
resistance protein {Pisum sativum}, partial (88%)
Length = 1107
Score = 183 bits (465), Expect(2) = 3e-64
Identities = 94/220 (42%), Positives = 141/220 (63%), Gaps = 3/220 (1%)
Frame = +3
Query: 5 KIKEHFELKAWVYVSESFDVVGLTKAILKSFNSSA-DGEDLNLLQHQLQYMLMGKKYLLV 63
++++HF+LKAWV VSE FD++ +TK++L+S S+ D ++L++L+ +L+ + K++L V
Sbjct: 12 EVQQHFDLKAWVCVSEDFDIMRVTKSLLESVTSTTWDSKNLDVLRVELKKISREKRFLFV 191
Query: 64 LDDIWNGDAERWELLLLPFNHGSFGSKIVVTTREKEVADNVLKSTELFDLQQLDKSNCWS 123
DD+WN + W L PF +G GS +++TTR+++VA+ V + + L+ L +CWS
Sbjct: 192 FDDLWNDNYNDWNELASPFINGKPGSMVIITTRQQKVAE-VAHTFPIHKLKLLSNEDCWS 368
Query: 124 LFVTHAFQGKSVSEYPN--LESVGRKIVEKCGGLPLAIKSLGQLLRKTFSEHEWINILET 181
L HA N LE GRKI KCGGLP+A K++G LLR EW +IL +
Sbjct: 369 LLSKHALGSDEFHHSSNTALEETGRKIARKCGGLPIAAKTIGGLLRSKVDITEWTSILNS 548
Query: 182 DMWRLSKVDHNVNSVLRLSYHNLPSNLKRCFSYCSIFPKG 221
++W L + N+ L LSY LPS+LKRCF+YCSIF KG
Sbjct: 549 NIWNLR--NDNILPALHLSYQYLPSHLKRCFAYCSIFSKG 662
Score = 81.6 bits (200), Expect(2) = 3e-64
Identities = 55/155 (35%), Positives = 79/155 (50%), Gaps = 5/155 (3%)
Frame = +1
Query: 218 FPKGHKFKKDELIMLWMAEGLLKCCGSNRSEEEFGNESFADLVSISFFQQSFDEIYDTYE 277
FPK + + EL++LWMAEG L C + EE G++ FA+L+S S QQ D+ D E
Sbjct: 652 FPKDYPLDRKELVLLWMAEGFLDCSQRGKKMEELGDDCFAELLSRSLIQQLSDD--DRGE 825
Query: 278 HYVMHDLVNDLTKSVSGEFSIQIEDARVERSVERTRHIWFSLQSNSV----DKLLELTC- 332
+VMHDLVNDL VSG+ ++E + E RH ++ ++ + +KL C
Sbjct: 826 KFVMHDLVNDLATFVSGKSCCRLECGDIP---ENVRHFSYNQENYDIFMKFEKLHNFKCL 996
Query: 333 EGLHSLILEGTRAMLISNNVQQDLFSRLNFLRMLS 367
+ L R +S V DL LR+LS
Sbjct: 997 RSFLFICLMTWRDNYLSFKVVNDLLPSHKRLRVLS 1101
>BF635265 similar to GP|4234955|gb|A NBS-LRR-like protein cD8 {Phaseolus
vulgaris}, partial (3%)
Length = 673
Score = 238 bits (607), Expect = 8e-63
Identities = 121/207 (58%), Positives = 149/207 (71%)
Frame = +1
Query: 524 GREEMDESMAESNVSVLEALQPNRNLKRLTISKYKGNSFPNWIRGYHLPNLVSLNLQFCG 583
G + MD+ E+ SVLEALQPN NL LTI Y+G SFPNW+ HLPNLVSL L C
Sbjct: 55 GEKWMDQ*-XEAQASVLEALQPNINLTSLTIKDYRGGSFPNWLGDRHLPNLVSLELLGCK 231
Query: 584 LCSLLPPLGTLPFLKMLSISDCDGIKIIGEEFYDSSSINVLFRSLEVLKFEKMNNWEEWL 643
+ S LPPLG P LK SIS CDGI+IIG EF +S +V FRSLE L+FE M W+EWL
Sbjct: 232 IHSQLPPLGQFPSLKKCSISSCDGIEIIGTEFLGYNSSDVPFRSLETLRFENMAEWKEWL 411
Query: 644 CLEGFPLLKELYIRECPKLKMSLPQHLPSLQKLFINDCKMLEASIPNGDNIIDLDIKRCD 703
CLEGFPLL++L I+ CPKLK +LPQHLPSLQKL I DC+ L ASIP NI +L++KRCD
Sbjct: 412 CLEGFPLLQKLCIKHCPKLKSALPQHLPSLQKLEIIDCQELAASIPKAANITELELKRCD 591
Query: 704 RILVNELPTSLKKLFILENRYTEFSVE 730
IL+NELP+ LK++ + + + ++E
Sbjct: 592 DILINELPSKLKRIILCGTQVIQSTLE 672
>TC83923 weakly similar to GP|14348622|gb|AAK61318.1 NBS-LRR resistance-like
protein J78 {Phaseolus vulgaris}, partial (2%)
Length = 551
Score = 214 bits (544), Expect(2) = 7e-61
Identities = 105/159 (66%), Positives = 125/159 (78%)
Frame = +1
Query: 552 LTISKYKGNSFPNWIRGYHLPNLVSLNLQFCGLCSLLPPLGTLPFLKMLSISDCDGIKII 611
L+IS+Y+GN FPNWIRG HLPNLVSL ++ CGLCS LPPLG LP L+ LSIS+C IKII
Sbjct: 73 LSISQYRGNRFPNWIRGCHLPNLVSLQMRHCGLCSHLPPLGQLPSLRELSISNCKRIKII 252
Query: 612 GEEFYDSSSINVLFRSLEVLKFEKMNNWEEWLCLEGFPLLKELYIRECPKLKMSLPQHLP 671
GEE Y ++S FRSLEVL+F++M N EEWLC EGF LKEL I++CPKLK +LPQHLP
Sbjct: 253 GEELYGNNSKIDAFRSLEVLEFQRMENLEEWLCHEGFLSLKELTIKDCPKLKRALPQHLP 432
Query: 672 SLQKLFINDCKMLEASIPNGDNIIDLDIKRCDRILVNEL 710
SLQKL I +C LEAS+P GDNI+ L +K CD L+ EL
Sbjct: 433 SLQKLSIINCNKLEASMPEGDNILKLCLKGCDSXLIKEL 549
Score = 39.7 bits (91), Expect(2) = 7e-61
Identities = 19/27 (70%), Positives = 22/27 (81%)
Frame = +2
Query: 529 DESMAESNVSVLEALQPNRNLKRLTIS 555
D+S+ ESNVSVLEALQPNR+LK S
Sbjct: 2 DDSIVESNVSVLEALQPNRSLKEALAS 82
Score = 33.1 bits (74), Expect = 0.52
Identities = 23/69 (33%), Positives = 34/69 (48%), Gaps = 6/69 (8%)
Frame = +1
Query: 855 PTLSYLNLNNCSKLRIM------NNEGFLHLKSLEFLYIINCPSLERLPEEALPNSLYSL 908
P+L L+++NC +++I+ NN +SLE L +LE SL L
Sbjct: 202 PSLRELSISNCKRIKIIGEELYGNNSKIDAFRSLEVLEFQRMENLEEWLCHEGFLSLKEL 381
Query: 909 WIKDCPLIK 917
IKDCP +K
Sbjct: 382 TIKDCPKLK 408
>TC81427 weakly similar to GP|11994216|dbj|BAB01338. disease resistance
comples protein {Arabidopsis thaliana}, partial (6%)
Length = 1253
Score = 226 bits (577), Expect = 2e-59
Identities = 140/346 (40%), Positives = 188/346 (53%), Gaps = 5/346 (1%)
Frame = +3
Query: 363 LRMLSFRGCGLLELVDEISNLKLLRYLDLSYTWIEILPDTICMLHNLQTLLLEGCCELTE 422
LR+ S + L I +L LRYLDLS T I LPD+IC L+NL LLL GC +LT
Sbjct: 96 LRVFSLSEYPITLLPSSIGHLLHLRYLDLSRTPITSLPDSICNLYNLXALLLVGCADLTL 275
Query: 423 LPSNFSKLVNLRHLKLPSHNGRPCIKTMPKHTGKLNNLQSLSYFIVEEQNVSDLKELAKL 482
LP+ SKL+NLR L + IK MP + GKL +LQSL F+V S++ EL ++
Sbjct: 276 LPTKTSKLINLRQLDISGSG----IKKMPTNLGKLKSLQSLPRFVVSNDGGSNVGELGEM 443
Query: 483 NHLHGAIDIEGLGNVSDLADSATVNLKDTKYLEELHMKFDGGREEMDESMAESNVSVLEA 542
L G++ I L NV ++ LK KYL E+ K+ ES + +
Sbjct: 444 LELRGSLSIVNLENVLLKEXASNAGLKRKKYLHEVEFKWTTPTHSQ-----ESENIIFDM 608
Query: 543 LQPNRNLKRLTISKYKGNSFPNWIRGYHLPNLVSLNLQFCGLCSLLPPLGTLPFLKMLSI 602
L P+RNLKRL I+ + G FPNW+ ++SL L CG C LP LG L L+ + I
Sbjct: 609 LXPHRNLKRLKINNFGGEKFPNWLGSNSGSTMMSLYLDECGNCLSLPSLGQLSNLREIYI 788
Query: 603 SDCDGIKIIGEEFYDSSSINVLFRSLEVLKFEKMNNWEEWLC-----LEGFPLLKELYIR 657
+ ++ +G EFY + F SL ++KF+ M NWEEW EGF LL+ELYI
Sbjct: 789 TSVTRLQKVGPEFYGNGF--EAFSSLRIIKFKDMLNWEEWSVNNQSGSEGFTLLQELYIE 962
Query: 658 ECPKLKMSLPQHLPSLQKLFINDCKMLEASIPNGDNIIDLDIKRCD 703
CPKL LP +LPSL KL I C+ L ++P + +L I C+
Sbjct: 963 NCPKLIGKLPGNLPSLDKLVITSCQTLSDTMPCVPRLRELKISGCE 1100
Score = 40.4 bits (93), Expect = 0.003
Identities = 30/105 (28%), Positives = 46/105 (43%)
Frame = +3
Query: 809 LLSLTITNCPKLIASRQEWGLKSLKYFFVCDDFENVESFPKESLLPPTLSYLNLNNCSKL 868
L L I NCPKLI + L SL + ++ P P L L ++ C
Sbjct: 942 LQELYIENCPKLIG-KLPGNLPSLDKLVITSCQTLSDTMP----CVPRLRELKISGCEAF 1106
Query: 869 RIMNNEGFLHLKSLEFLYIINCPSLERLPEEALPNSLYSLWIKDC 913
++ + L+ + I NCPSL +P + + +L SL + DC
Sbjct: 1107 VSLSEQMMKCNDCLQTMAISNCPSLVSIPMDCVSRTLKSLKVSDC 1241
Score = 32.0 bits (71), Expect = 1.2
Identities = 39/153 (25%), Positives = 56/153 (36%)
Frame = +3
Query: 761 NSLGELSITRWCSSSLSFSLHLFTNLYSLWFVDCPNLDSFPEGGLPCNLLSLTITNCPKL 820
N+ G W S+ + + SL+ +C N S P G NL + IT+ +L
Sbjct: 645 NNFGGEKFPNWLGSNSG------STMMSLYLDECGNCLSLPSLGQLSNLREIYITSVTRL 806
Query: 821 IASRQEWGLKSLKYFFVCDDFENVESFPKESLLPPTLSYLNLNNCSKLRIMNNEGFLHLK 880
E F + FE S + + ++ N + + N G
Sbjct: 807 QKVGPE---------FYGNGFEAFSSLR-------IIKFKDMLNWEEWSVNNQSGSEGFT 938
Query: 881 SLEFLYIINCPSLERLPEEALPNSLYSLWIKDC 913
L+ LYI NCP L LP SL L I C
Sbjct: 939 LLQELYIENCPKLIGKLPGNLP-SLDKLVITSC 1034
>BF518621 weakly similar to GP|11994216|db disease resistance comples protein
{Arabidopsis thaliana}, partial (2%)
Length = 457
Score = 206 bits (524), Expect = 3e-53
Identities = 102/150 (68%), Positives = 118/150 (78%), Gaps = 3/150 (2%)
Frame = +3
Query: 781 HLFTNLYSLWFVDCPNLDSFPEGGLPCNLLSLTITNCPKLIASRQEWGL---KSLKYFFV 837
HLFTNL SL DCP L+SFPEGGLP NL L I NCPKLIASR++W L SLKYF V
Sbjct: 3 HLFTNLDSLKLRDCPELESFPEGGLPSNLRKLEINNCPKLIASREDWDLFQLNSLKYFIV 182
Query: 838 CDDFENVESFPKESLLPPTLSYLNLNNCSKLRIMNNEGFLHLKSLEFLYIINCPSLERLP 897
CDDF+ +ESFP+ESLLPPTL L L+ CSKLRIMN +G LHLKSL+ LYI CPSLERLP
Sbjct: 183 CDDFKTMESFPEESLLPPTLHTLFLDKCSKLRIMNYKGLLHLKSLKVLYIGRCPSLERLP 362
Query: 898 EEALPNSLYSLWIKDCPLIKVKYQKEGGEQ 927
EE +PNSL L I DCPL++ +Y+KEGG++
Sbjct: 363 EEGIPNSLSRLVISDCPLLEQQYRKEGGDR 452
Score = 50.1 bits (118), Expect = 4e-06
Identities = 53/174 (30%), Positives = 76/174 (43%), Gaps = 1/174 (0%)
Frame = +3
Query: 648 FPLLKELYIRECPKLKMSLPQHLPS-LQKLFINDCKMLEASIPNGDNIIDLDIKRCDRIL 706
F L L +R+CP+L+ LPS L+KL IN+C L AS + D + L+
Sbjct: 9 FTNLDSLKLRDCPELESFPEGGLPSNLRKLEINNCPKLIASREDWD-LFQLN-------- 161
Query: 707 VNELPTSLKKLFILENRYTEFSVEQIFVNSTILEVLELDLNGSLKCPTLDLCCYNSLGEL 766
SLK + ++ T S + + L L LD KC L + Y L
Sbjct: 162 ------SLKYFIVCDDFKTMESFPEESLLPPTLHTLFLD-----KCSKLRIMNYKGL--- 299
Query: 767 SITRWCSSSLSFSLHLFTNLYSLWFVDCPNLDSFPEGGLPCNLLSLTITNCPKL 820
LHL +L L+ CP+L+ PE G+P +L L I++CP L
Sbjct: 300 -------------LHL-KSLKVLYIGRCPSLERLPEEGIPNSLSRLVISDCPLL 419
Score = 38.1 bits (87), Expect = 0.016
Identities = 40/140 (28%), Positives = 61/140 (43%), Gaps = 27/140 (19%)
Frame = +3
Query: 573 NLVSLNLQFCGLCSLLPPLGTLPFLKMLSISDCDGIKIIGEEFYDSSSINVL-------- 624
NL SL L+ C P G L+ L I++C + I E +D +N L
Sbjct: 15 NLDSLKLRDCPELESFPEGGLPSNLRKLEINNCPKL-IASREDWDLFQLNSLKYFIVCDD 191
Query: 625 FRSLEVLKFE------------------KMNNWEEWLCLEGFPLLKELYIRECPKLKMSL 666
F+++E E ++ N++ L L+ LK LYI CP L+
Sbjct: 192 FKTMESFPEESLLPPTLHTLFLDKCSKLRIMNYKGLLHLKS---LKVLYIGRCPSLERLP 362
Query: 667 PQHLP-SLQKLFINDCKMLE 685
+ +P SL +L I+DC +LE
Sbjct: 363 EEGIPNSLSRLVISDCPLLE 422
>BG645249
Length = 434
Score = 191 bits (486), Expect = 9e-49
Identities = 99/143 (69%), Positives = 108/143 (75%), Gaps = 3/143 (2%)
Frame = +1
Query: 759 CYNSLGELSITRWCSSSLSFSLHLFTNLYSLWFVDCPNLDSFPEGGLPCNLLSLTITNCP 818
CYNSL LSI W SSSL LHLFT+L SL+ DCP L+SFP GGLP NL L I NCP
Sbjct: 4 CYNSLQRLSIEGWGSSSLPLELHLFTSLRSLYLDDCPELESFPMGGLPSNLRDLRIHNCP 183
Query: 819 KLIASRQEWGL---KSLKYFFVCDDFENVESFPKESLLPPTLSYLNLNNCSKLRIMNNEG 875
KLI SR+EWGL SLK+F V D+FENVESFP+E+LLPPTL L L NCSKLR MN +G
Sbjct: 184 KLIGSREEWGLFQLNSLKWFSVSDEFENVESFPEENLLPPTLKDLYLINCSKLRKMNKKG 363
Query: 876 FLHLKSLEFLYIINCPSLERLPE 898
FLHLKSL LYI NCPSLE LPE
Sbjct: 364 FLHLKSLNKLYIRNCPSLESLPE 432
Score = 36.6 bits (83), Expect = 0.047
Identities = 19/39 (48%), Positives = 24/39 (60%), Gaps = 1/39 (2%)
Frame = +1
Query: 877 LHL-KSLEFLYIINCPSLERLPEEALPNSLYSLWIKDCP 914
LHL SL LY+ +CP LE P LP++L L I +CP
Sbjct: 67 LHLFTSLRSLYLDDCPELESFPMGGLPSNLRDLRIHNCP 183
Score = 33.9 bits (76), Expect = 0.30
Identities = 46/178 (25%), Positives = 72/178 (39%)
Frame = +1
Query: 625 FRSLEVLKFEKMNNWEEWLCLEGFPLLKELYIRECPKLKMSLPQHLPSLQKLFINDCKML 684
+ SL+ L E + L L F L+ LY+ +CP+L+ LPS
Sbjct: 7 YNSLQRLSIEGWGSSSLPLELHLFTSLRSLYLDDCPELESFPMGGLPS------------ 150
Query: 685 EASIPNGDNIIDLDIKRCDRILVNELPTSLKKLFILENRYTEFSVEQIFVNSTILEVLEL 744
N+ DL I C +++ + L +L N FSV F N +E
Sbjct: 151 --------NLRDLRIHNCPKLIGSREEWGLFQL----NSLKWFSVSDEFEN------VES 276
Query: 745 DLNGSLKCPTLDLCCYNSLGELSITRWCSSSLSFSLHLFTNLYSLWFVDCPNLDSFPE 802
+L PTL L ++ ++ + LHL +L L+ +CP+L+S PE
Sbjct: 277 FPEENLLPPTL-----KDLYLINCSKLRKMNKKGFLHL-KSLNKLYIRNCPSLESLPE 432
>TC87193 similar to GP|21616922|gb|AAM66423.1 NBS-LRR protein {Oryza sativa
(japonica cultivar-group)}, partial (26%)
Length = 3119
Score = 134 bits (337), Expect(2) = 5e-47
Identities = 82/185 (44%), Positives = 106/185 (56%), Gaps = 5/185 (2%)
Frame = +3
Query: 1 YNDNKIKEHFELKAWVYVSESFDVVGLTKAILKSFNSSADGED-----LNLLQHQLQYML 55
Y D ++K FE WV VS++FD K ILK+ +S ED L LQ LQ L
Sbjct: 708 YKDGEVKNLFEKHMWVCVSDNFDF----KTILKNMVASLTKEDVVNKTLQELQSMLQVNL 875
Query: 56 MGKKYLLVLDDIWNGDAERWELLLLPFNHGSFGSKIVVTTREKEVADNVLKSTELFDLQQ 115
GK+YLLVLDD+WN E+W+ L G+ GSK+V+TT K VAD + S + L+
Sbjct: 876 TGKRYLLVLDDVWNESFEKWDQLRPYLMCGAQGSKVVMTTCSKIVADRMGVSDQHV-LRG 1052
Query: 116 LDKSNCWSLFVTHAFQGKSVSEYPNLESVGRKIVEKCGGLPLAIKSLGQLLRKTFSEHEW 175
L W LF F +V LES+G+KI EKC G+PLAI+SLG +LR E EW
Sbjct: 1053LTPEKSWVLFKNIVFGDVTVGVNQPLESIGKKIAEKCKGVPLAIRSLGGILRSESKESEW 1232
Query: 176 INILE 180
IN+L+
Sbjct: 1233INVLQ 1247
Score = 73.2 bits (178), Expect(2) = 5e-47
Identities = 32/73 (43%), Positives = 48/73 (64%)
Frame = +1
Query: 196 VLRLSYHNLPSNLKRCFSYCSIFPKGHKFKKDELIMLWMAEGLLKCCGSNRSEEEFGNES 255
VL+LSY NL ++CF+YCS+FP+ +F+KDELI +WMA+G L C + ++ GN+
Sbjct: 1297 VLKLSYQNLSPQQRQCFAYCSLFPQDWEFEKDELIQMWMAQGYLGCSVEKQCMKDVGNQF 1476
Query: 256 FADLVSISFFQQS 268
+ SFFQ +
Sbjct: 1477 VKIFLKNSFFQDA 1515
Score = 43.5 bits (101), Expect = 4e-04
Identities = 31/94 (32%), Positives = 49/94 (51%), Gaps = 4/94 (4%)
Frame = +2
Query: 550 KRLTISKYKGNSFPNWIRGYHLPNLVSLNLQFCGLCSLLPPLGTLPFLKMLSISDCDGIK 609
++L+I + KG S NW+ + N++ L L C LPPL LPFLK L + + ++
Sbjct: 2054 RKLSIQQPKGLSLSNWLSP--VTNIIELCLYNCQDLRYLPPLERLPFLKSLDLRWLNQLE 2227
Query: 610 IIGEEFYDSSSIN-VLFRSLEVLKF---EKMNNW 639
I +Y+ ++ F SLE+L F K+ W
Sbjct: 2228 YI---YYEEPILHESFFPSLEILNFFECWKLKGW 2320
Score = 42.0 bits (97), Expect = 0.001
Identities = 33/84 (39%), Positives = 44/84 (52%), Gaps = 1/84 (1%)
Frame = +2
Query: 356 LFSRLNFLRMLSFRGCGLLELVDEISNLKLLRYLDL-SYTWIEILPDTICMLHNLQTLLL 414
+F L +LR L C + V I LK LR+LDL +Y E L +I L LQT+ L
Sbjct: 1772 VFRNLKYLRFLKMESCSR-QGVGFIEKLKHLRHLDLRNYDSGESLSKSIGNLVCLQTIKL 1948
Query: 415 EGCCELTELPSNFSKLVNLRHLKL 438
+ + P SKL+NLRHL +
Sbjct: 1949 ND--SVVDSPEVVSKLINLRHLTI 2014
Score = 40.4 bits (93), Expect = 0.003
Identities = 20/61 (32%), Positives = 34/61 (54%)
Frame = +1
Query: 378 DEISNLKLLRYLDLSYTWIEILPDTICMLHNLQTLLLEGCCELTELPSNFSKLVNLRHLK 437
D++ L L+ + + ++ LPD IC + +LQ L + GC L +LP +L NL L+
Sbjct: 2698 DDLICLPSLQKIQIRTCRLKALPDWICNISSLQHLAVYGCESLVDLPEGMPRLTNLHTLE 2877
Query: 438 L 438
+
Sbjct: 2878 I 2880
>BG587581 weakly similar to GP|11994216|db disease resistance comples protein
{Arabidopsis thaliana}, partial (5%)
Length = 641
Score = 178 bits (451), Expect(2) = 1e-44
Identities = 108/194 (55%), Positives = 132/194 (67%), Gaps = 6/194 (3%)
Frame = +2
Query: 249 EEFGNESFADLVSISFFQQSFDEIYDTYEHYVMHDLVNDLTKSVSGEFSIQIEDARVERS 308
++FGNE F DLVSISFFQQS D +VMHDLVNDL KS+ GEF + I+ + +
Sbjct: 14 QDFGNELFVDLVSISFFQQSTDGS----TKFVMHDLVNDLAKSMVGEFCLAIQGDKEKDV 181
Query: 309 VERTRHIWFS-LQSNSVDKLLE--LTCEGLHSLILEGTRAML---ISNNVQQDLFSRLNF 362
ERTRHI S Q +K+ + +GL SL++ + ISN +QQDLFS+L
Sbjct: 182 TERTRHISCSQFQRKDANKMTQHIYKTKGLRSLLVYLNSDVFHQNISNAIQQDLFSKLKC 361
Query: 363 LRMLSFRGCGLLELVDEISNLKLLRYLDLSYTWIEILPDTICMLHNLQTLLLEGCCELTE 422
LRMLS GC L +L DE+SNLKLLRYLDLSYT IE LPD+IC L+NLQTLLL+ C LTE
Sbjct: 362 LRMLSLNGCILPKLDDEVSNLKLLRYLDLSYTRIESLPDSICNLYNLQTLLLKN-CPLTE 538
Query: 423 LPSNFSKLVNLRHL 436
LPS+F KL +L HL
Sbjct: 539 LPSDFYKLSHLHHL 580
Score = 21.6 bits (44), Expect(2) = 1e-44
Identities = 10/17 (58%), Positives = 11/17 (63%)
Frame = +3
Query: 444 RPCIKTMPKHTGKLNNL 460
R IK MPK GKL +L
Sbjct: 591 RTHIKMMPKDIGKLTHL 641
>BF645813 weakly similar to GP|14348622|gb NBS-LRR resistance-like protein
J78 {Phaseolus vulgaris}, partial (8%)
Length = 620
Score = 163 bits (413), Expect(2) = 3e-42
Identities = 87/148 (58%), Positives = 109/148 (72%)
Frame = +3
Query: 347 LISNNVQQDLFSRLNFLRMLSFRGCGLLELVDEISNLKLLRYLDLSYTWIEILPDTICML 406
+ISNN+Q DLFS+L +LR+LSF GC L EL EI NLKLLRYL+L+ + IE LPD+IC L
Sbjct: 84 IISNNLQHDLFSKLKYLRLLSFGGCKLKELSGEIGNLKLLRYLNLTKSLIERLPDSICKL 263
Query: 407 HNLQTLLLEGCCELTELPSNFSKLVNLRHLKLPSHNGRPCIKTMPKHTGKLNNLQSLSYF 466
+ L+TL+LE C ELT LPS F KLV+LRHL L N IK MPK G LN+LQ+LS+F
Sbjct: 264 YKLETLILERCSELTILPSKFYKLVSLRHLNLKGCN----IKKMPKQMGSLNHLQTLSHF 431
Query: 467 IVEEQNVSDLKELAKLNHLHGAIDIEGL 494
+V E+N S+++EL LN L G + I GL
Sbjct: 432 VVGEENGSNIQELGNLNRLQGKLCISGL 515
Score = 27.7 bits (60), Expect(2) = 3e-42
Identities = 11/20 (55%), Positives = 16/20 (80%)
Frame = +1
Query: 502 DSATVNLKDTKYLEELHMKF 521
D+A NLKD K++ EL+MK+
Sbjct: 538 DAARANLKDKKHVXELNMKY 597
>TC83046 weakly similar to GP|10280097|emb|CAC09980. nucleotide sequence of
the I-2 resistance gene {Fusarium oxysporum}, partial
(8%)
Length = 884
Score = 166 bits (421), Expect = 3e-41
Identities = 87/166 (52%), Positives = 110/166 (65%)
Frame = +1
Query: 136 SEYPNLESVGRKIVEKCGGLPLAIKSLGQLLRKTFSEHEWINILETDMWRLSKVDHNVNS 195
S+ PNLE +GRKI +KCGGLP+A K+LG +LR EW IL +D+W L + N+
Sbjct: 1 SKCPNLEEIGRKIAKKCGGLPIAAKTLGGILRSKVDAKEWSTILNSDIWNLP--NDNILP 174
Query: 196 VLRLSYHNLPSNLKRCFSYCSIFPKGHKFKKDELIMLWMAEGLLKCCGSNRSEEEFGNES 255
LRLSY LPS+LKRCF+YCSIFPK K ELI+LWMAEG L+ N++ EE G++
Sbjct: 175 ALRLSYQYLPSHLKRCFAYCSIFPKDFSLDKKELILLWMAEGFLEHSQCNKTAEEVGHDY 354
Query: 256 FADLVSISFFQQSFDEIYDTYEHYVMHDLVNDLTKSVSGEFSIQIE 301
F +L+S S QQS D D E +VMHDLVNDL VSG ++E
Sbjct: 355 FIELLSRSLIQQSND---DGKEKFVMHDLVNDLALVVSGTSCFRLE 483
>BF639307 similar to GP|11994216|db disease resistance comples protein
{Arabidopsis thaliana}, partial (1%)
Length = 637
Score = 159 bits (401), Expect = 6e-39
Identities = 79/111 (71%), Positives = 90/111 (80%), Gaps = 1/111 (0%)
Frame = +1
Query: 607 GIKIIGEEFYDSSSINVLFRSLEVLKFEKMNNWEEWLCLEGFPLLKELYIRECPKLKMS- 665
GIKIIGEE + ++ +V F SLEVLK E M NWEEW C EGFPLLKEL IR CPKLK +
Sbjct: 91 GIKIIGEEIHGNNLTHVPFLSLEVLKLEDMVNWEEWFCREGFPLLKELTIRNCPKLKRAL 270
Query: 666 LPQHLPSLQKLFINDCKMLEASIPNGDNIIDLDIKRCDRILVNELPTSLKK 716
LPQHLPSLQKL + DCK LE S+P GDNII+L ++RCDRILVNELPTSLK+
Sbjct: 271 LPQHLPSLQKLELCDCKQLEVSVPKGDNIIELKMQRCDRILVNELPTSLKR 423
Score = 101 bits (252), Expect = 1e-21
Identities = 86/231 (37%), Positives = 110/231 (47%), Gaps = 27/231 (11%)
Frame = +2
Query: 583 GLCSLLPPLGTLPFLKMLSISDCDGIKIIGEEFYDS-------------SSINVLFRSLE 629
GLCS LPPLG LP LK E FY ++I +FRS
Sbjct: 17 GLCSHLPPLGQLPSLK--------------EPFYFKM*PE*RSLVKRFMATI*RMFRSCP 154
Query: 630 VLKFEKMNNWEEW------LCLEGFPLLKELYIRECPKLKMSLPQHLPSLQK----LFIN 679
+ K NW W ++GF LK L P K + L K + +N
Sbjct: 155 L----KF*NWRIWSIGRNGFVVKGFLCLKSLL*EIAPN*KGPCCLNTFLLYKNWNFVIVN 322
Query: 680 DCKMLEASIPNGDNIIDLDIKRCDRIL----VNELPTSLKKLFILENRYTEFSVEQIFVN 735
+ +I RC ++ +N P + K + + +NRYTEFSV+Q +N
Sbjct: 323 SWRY------QFPRVIIS*S*RCRDVIEFW*MNCRP-A*KGVLLCDNRYTEFSVDQNLIN 481
Query: 736 STILEVLELDLNGSLKCPTLDLCCYNSLGELSITRWCSSSLSFSLHLFTNL 786
LE LELD +GS+KCP+LDLCCYNSL LSI+ W SSSL FSLHLFT L
Sbjct: 482 FPFLEELELDWSGSVKCPSLDLCCYNSLSTLSISGWGSSSLPFSLHLFTIL 634
Score = 29.6 bits (65), Expect = 5.7
Identities = 31/93 (33%), Positives = 40/93 (42%), Gaps = 1/93 (1%)
Frame = +1
Query: 783 FTNLYSLWFVDCPNLDS-FPEGGLPCNLLSLTITNCPKLIASRQEWGLKSLKYFFVCDDF 841
F +L L D N + F G P L LTI NCPKL + L SL+ +CD
Sbjct: 145 FLSLEVLKLEDMVNWEEWFCREGFPL-LKELTIRNCPKLKRALLPQHLPSLQKLELCDCK 321
Query: 842 ENVESFPKESLLPPTLSYLNLNNCSKLRIMNNE 874
+ S PK + L + C RI+ NE
Sbjct: 322 QLEVSVPKGD----NIIELKMQRCD--RILVNE 402
>BM815640 weakly similar to GP|14348619|gb NBS-LRR resistance-like protein
J71 {Phaseolus vulgaris}, partial (3%)
Length = 715
Score = 103 bits (257), Expect(2) = 4e-38
Identities = 51/80 (63%), Positives = 61/80 (75%), Gaps = 5/80 (6%)
Frame = -1
Query: 783 FTNLYSLWFVDCPNLDSFPEGGLPCNLLSLTITNCPKLIASRQEWG-----LKSLKYFFV 837
FT+L +L+ DCP L+SFP GGLP NL L+I NCPKLI SR+EWG L SLKYF +
Sbjct: 469 FTSLDTLYLYDCPELESFPMGGLPPNLSDLSICNCPKLIGSREEWGLFQFQLNSLKYFTI 290
Query: 838 CDDFENVESFPKESLLPPTL 857
DDFENVESF +E+LLPP+L
Sbjct: 289 DDDFENVESFLEENLLPPSL 230
Score = 73.9 bits (180), Expect(2) = 4e-38
Identities = 42/67 (62%), Positives = 52/67 (76%), Gaps = 3/67 (4%)
Frame = -3
Query: 856 TLSYL-NLNNCSKLRIMNNEGFLHLKSLEFLYIINCPSLERLPE-EALPNSLYSLWIK-D 912
T+S+L L N SKLRIMN +GFLHLKSL+ LYI +CP+LE L E E LPNSL +LWI+ +
Sbjct: 239 TISFLFGLKNFSKLRIMNKKGFLHLKSLDCLYIEDCPTLESLTEKEDLPNSLTTLWIEPN 60
Query: 913 CPLIKVK 919
C +IK K
Sbjct: 59 CRIIKEK 39
Score = 35.0 bits (79), Expect = 0.14
Identities = 16/34 (47%), Positives = 21/34 (61%)
Frame = -1
Query: 881 SLEFLYIINCPSLERLPEEALPNSLYSLWIKDCP 914
SL+ LY+ +CP LE P LP +L L I +CP
Sbjct: 463 SLDTLYLYDCPELESFPMGGLPPNLSDLSICNCP 362
Score = 32.7 bits (73), Expect = 0.67
Identities = 31/76 (40%), Positives = 37/76 (47%), Gaps = 7/76 (9%)
Frame = -1
Query: 840 DFENVESFPKESLLPPTLSYLNLNNCSKLRIMNNEGFL---HLKSLEFLYI----INCPS 892
D +ESFP L PP LS L++ NC KL E L L SL++ I N S
Sbjct: 439 DCPELESFPMGGL-PPNLSDLSICNCPKLIGSREEWGLFQFQLNSLKYFTIDDDFENVES 263
Query: 893 LERLPEEALPNSLYSL 908
L E LP SL+SL
Sbjct: 262 F--LEENLLPPSLFSL 221
>BG587633 similar to GP|20385442|gb| resistance gene analog {Vitis vinifera},
partial (22%)
Length = 677
Score = 154 bits (388), Expect = 2e-37
Identities = 76/161 (47%), Positives = 105/161 (65%)
Frame = +3
Query: 141 LESVGRKIVEKCGGLPLAIKSLGQLLRKTFSEHEWINILETDMWRLSKVDHNVNSVLRLS 200
LE++GRK KCGGLP+A K+LG LLR EW +IL +D+W LS + N+ L LS
Sbjct: 27 LEAIGRKTARKCGGLPIAAKTLGGLLRSKVEITEWTSILNSDIWNLS--NDNILPALHLS 200
Query: 201 YHNLPSNLKRCFSYCSIFPKGHKFKKDELIMLWMAEGLLKCCGSNRSEEEFGNESFADLV 260
Y LP +LKRCF+YCSIFPK + + +L++LWMAEG L C ++ EE G++ FA+L+
Sbjct: 201 YQYLPCHLKRCFAYCSIFPKDYPLDRKQLVLLWMAEGFLDCSHGGKAMEELGDDCFAELL 380
Query: 261 SISFFQQSFDEIYDTYEHYVMHDLVNDLTKSVSGEFSIQIE 301
S S QQ ++ E +VMHDLVNDL +SG+ ++
Sbjct: 381 SRSLIQQLSNDARG--EKFVMHDLVNDLATVISGQSCFNLD 497
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.321 0.139 0.420
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 35,090,278
Number of Sequences: 36976
Number of extensions: 578574
Number of successful extensions: 3719
Number of sequences better than 10.0: 229
Number of HSP's better than 10.0 without gapping: 3352
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 3567
length of query: 946
length of database: 9,014,727
effective HSP length: 105
effective length of query: 841
effective length of database: 5,132,247
effective search space: 4316219727
effective search space used: 4316219727
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 63 (28.9 bits)
Medicago: description of AC146704.8


