

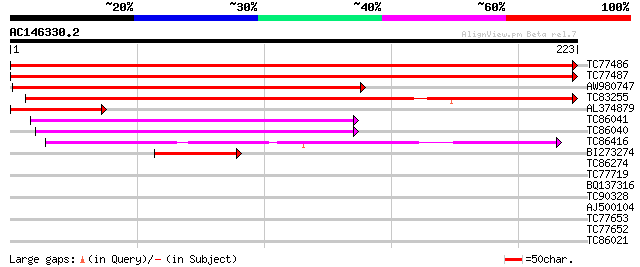
BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146330.2 + phase: 0
(223 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC77486 homologue to SP|O82531|PSB1_PETHY Proteasome subunit bet... 452 e-128
TC77487 homologue to SP|O82531|PSB1_PETHY Proteasome subunit bet... 447 e-126
AW980747 similar to SP|O82531|PSB1 Proteasome subunit beta type ... 244 2e-65
TC83255 weakly similar to SP|Q9IB83|PS12_CARAU Proteasome subuni... 206 6e-54
AL374879 homologue to GP|600387|emb|C proteosome subunit {Arabid... 86 1e-17
TC86041 homologue to GP|17473525|gb|AAL38246.1 proteasome subuni... 78 2e-15
TC86040 homologue to GP|17473525|gb|AAL38246.1 proteasome subuni... 75 2e-14
TC86416 homologue to PIR|T04497|T04497 multicatalytic endopeptid... 71 4e-13
BI273274 similar to GP|14594933|emb putative beta6 proteasome su... 66 1e-11
TC86274 similar to PIR|T51986|T51986 multicatalytic endopeptidas... 37 0.006
TC77719 similar to GP|21593260|gb|AAM65209.1 unknown {Arabidopsi... 32 0.19
BQ137316 32 0.19
TC90328 similar to PIR|B96706|B96706 unknown protein 56300-5788... 29 1.3
AJ500104 weakly similar to PIR|T38282|S624 probable proteasome c... 28 3.6
TC77653 similar to GP|20334820|gb|AAM16166.1 At1g12050/F12F1_8 {... 28 3.6
TC77652 similar to GP|20334820|gb|AAM16166.1 At1g12050/F12F1_8 {... 28 3.6
TC86021 homologue to GP|19423939|gb|AAL87312.1 putative RNA heli... 27 6.2
>TC77486 homologue to SP|O82531|PSB1_PETHY Proteasome subunit beta type 1
(EC 3.4.25.1) (20S proteasome alpha subunit F), complete
Length = 1070
Score = 452 bits (1162), Expect = e-128
Identities = 223/223 (100%), Positives = 223/223 (100%)
Frame = +3
Query: 1 MTKQHANWSPYDNNGGSCVAIAGSDYCVIAADTRMSTGYNILTRDYSKISHLAEKCVMAS 60
MTKQHANWSPYDNNGGSCVAIAGSDYCVIAADTRMSTGYNILTRDYSKISHLAEKCVMAS
Sbjct: 117 MTKQHANWSPYDNNGGSCVAIAGSDYCVIAADTRMSTGYNILTRDYSKISHLAEKCVMAS 296
Query: 61 SGFQADVKALQKVLSARHLTYQHQHNKQMSCPAMAQLLSNTLYYKRFFPYYSFNVLGGLD 120
SGFQADVKALQKVLSARHLTYQHQHNKQMSCPAMAQLLSNTLYYKRFFPYYSFNVLGGLD
Sbjct: 297 SGFQADVKALQKVLSARHLTYQHQHNKQMSCPAMAQLLSNTLYYKRFFPYYSFNVLGGLD 476
Query: 121 SEGKGCVFTYDAVGSYERVGYSSQGSGSTLIMPFLDNQLKSPSPLLLPAQDAVTPLSEAE 180
SEGKGCVFTYDAVGSYERVGYSSQGSGSTLIMPFLDNQLKSPSPLLLPAQDAVTPLSEAE
Sbjct: 477 SEGKGCVFTYDAVGSYERVGYSSQGSGSTLIMPFLDNQLKSPSPLLLPAQDAVTPLSEAE 656
Query: 181 AVDLVKTVFASATERDIYTGDKVEIVILNAGGIHREFMDLRKD 223
AVDLVKTVFASATERDIYTGDKVEIVILNAGGIHREFMDLRKD
Sbjct: 657 AVDLVKTVFASATERDIYTGDKVEIVILNAGGIHREFMDLRKD 785
>TC77487 homologue to SP|O82531|PSB1_PETHY Proteasome subunit beta type 1
(EC 3.4.25.1) (20S proteasome alpha subunit F), complete
Length = 881
Score = 447 bits (1149), Expect = e-126
Identities = 220/223 (98%), Positives = 221/223 (98%)
Frame = +2
Query: 1 MTKQHANWSPYDNNGGSCVAIAGSDYCVIAADTRMSTGYNILTRDYSKISHLAEKCVMAS 60
MTKQHANWSPYDNNGGSCVAIAGSDYCVIAADTRMSTGYNILTRDYSKISHLAEKCVMAS
Sbjct: 35 MTKQHANWSPYDNNGGSCVAIAGSDYCVIAADTRMSTGYNILTRDYSKISHLAEKCVMAS 214
Query: 61 SGFQADVKALQKVLSARHLTYQHQHNKQMSCPAMAQLLSNTLYYKRFFPYYSFNVLGGLD 120
SGFQADVKALQKVLSARHLTYQHQHNKQMSCPAMAQLLSNTLYYKRFFPYYSFNVLGGLD
Sbjct: 215 SGFQADVKALQKVLSARHLTYQHQHNKQMSCPAMAQLLSNTLYYKRFFPYYSFNVLGGLD 394
Query: 121 SEGKGCVFTYDAVGSYERVGYSSQGSGSTLIMPFLDNQLKSPSPLLLPAQDAVTPLSEAE 180
SEGKGCVFTYDAVGSYERVGYSSQGSGSTLIMPFLDNQLKSPSPLLLPAQDAVTPLSE E
Sbjct: 395 SEGKGCVFTYDAVGSYERVGYSSQGSGSTLIMPFLDNQLKSPSPLLLPAQDAVTPLSETE 574
Query: 181 AVDLVKTVFASATERDIYTGDKVEIVILNAGGIHREFMDLRKD 223
AVDLVKTVFASATERDIYTGDKVEIVILNA GIHR+FMDLRKD
Sbjct: 575 AVDLVKTVFASATERDIYTGDKVEIVILNASGIHRDFMDLRKD 703
>AW980747 similar to SP|O82531|PSB1 Proteasome subunit beta type 1 (EC
3.4.25.1) (20S proteasome alpha subunit F), partial
(56%)
Length = 420
Score = 244 bits (622), Expect = 2e-65
Identities = 117/139 (84%), Positives = 124/139 (89%)
Frame = +2
Query: 2 TKQHANWSPYDNNGGSCVAIAGSDYCVIAADTRMSTGYNILTRDYSKISHLAEKCVMASS 61
TKQ A WSPYDN+GGSCVAIAGSDYCVIAADTRMSTGYNILTRDYSKISHLAEKCVMAS
Sbjct: 2 TKQDAIWSPYDNHGGSCVAIAGSDYCVIAADTRMSTGYNILTRDYSKISHLAEKCVMASI 181
Query: 62 GFQADVKALQKVLSARHLTYQHQHNKQMSCPAMAQLLSNTLYYKRFFPYYSFNVLGGLDS 121
FQ +++ L +V+ HL+Y HNKQMSCPAMA LLSNTLYYKRFFPYYSFNVLGGLDS
Sbjct: 182 DFQENMRCLHEVMLIDHLSYDGNHNKQMSCPAMAHLLSNTLYYKRFFPYYSFNVLGGLDS 361
Query: 122 EGKGCVFTYDAVGSYERVG 140
E KGCVFTYDAVGSYERVG
Sbjct: 362 ERKGCVFTYDAVGSYERVG 418
>TC83255 weakly similar to SP|Q9IB83|PS12_CARAU Proteasome subunit beta type
1-B (EC 3.4.25.1) (20S proteasome beta 6 subunit B)
(B6-B). [Goldfish], partial (91%)
Length = 796
Score = 206 bits (524), Expect = 6e-54
Identities = 109/218 (50%), Positives = 149/218 (68%), Gaps = 1/218 (0%)
Frame = +3
Query: 7 NWSPYDNNGGSCVAIAGSDYCVIAADTRMSTGYNILTRDYSKISHLAEKCVMASSGFQAD 66
N+ PY +NGG+ + IAG+D+C++A+DTR S+GY+I +R K L EK V+A++GF AD
Sbjct: 75 NFYPYADNGGTSLGIAGADFCLMASDTRKSSGYSIHSRYSPKAFKLTEKAVLATNGFAAD 254
Query: 67 VKALQKVLSARHLTYQHQHNKQMSCPAMAQLLSNTLYYKRFFPYYSFNVLGGLDSEGKGC 126
AL K L + Y+H H+K+MS PA+AQ+L+NTLYYKRFFPYY+FN+LGG+D +G+G
Sbjct: 255 GLALTKRLGQKLEWYKHAHDKEMSTPALAQMLANTLYYKRFFPYYTFNLLGGIDEQGRGV 434
Query: 127 VFTYDAVGSYERVGYSSQGSGSTLIMPFLDNQLKSPSPLLLPAQDA-VTPLSEAEAVDLV 185
++++D VGS + GS + LI PFLDNQ+ L Q A TPLS A+ +
Sbjct: 435 LYSFDPVGSMGTEICRAGGSSAALIQPFLDNQV-----ALKNQQGAQQTPLSLETAIRIT 599
Query: 186 KTVFASATERDIYTGDKVEIVILNAGGIHREFMDLRKD 223
K F SATERDIYTGD +EI I+ G E +DL+KD
Sbjct: 600 KDAFTSATERDIYTGDFLEIFIIRQEGTTIEMIDLKKD 713
>AL374879 homologue to GP|600387|emb|C proteosome subunit {Arabidopsis
thaliana}, partial (16%)
Length = 395
Score = 85.5 bits (210), Expect = 1e-17
Identities = 38/38 (100%), Positives = 38/38 (100%)
Frame = +1
Query: 1 MTKQHANWSPYDNNGGSCVAIAGSDYCVIAADTRMSTG 38
MTKQHANWSPYDNNGGSCVAIAGSDYCVIAADTRMSTG
Sbjct: 34 MTKQHANWSPYDNNGGSCVAIAGSDYCVIAADTRMSTG 147
>TC86041 homologue to GP|17473525|gb|AAL38246.1 proteasome subunit
{Arabidopsis thaliana}, complete
Length = 1090
Score = 78.2 bits (191), Expect = 2e-15
Identities = 42/129 (32%), Positives = 67/129 (51%)
Frame = +2
Query: 9 SPYDNNGGSCVAIAGSDYCVIAADTRMSTGYNILTRDYSKISHLAEKCVMASSGFQADVK 68
S ++ NG + VA+ G + IA+D R+ + D+ +IS + +K + SG D +
Sbjct: 101 SIFEYNGSALVAMVGKNCFAIASDRRLGVQLQTIATDFQRISKIHDKLFIGLSGLATDSQ 280
Query: 69 ALQKVLSARHLTYQHQHNKQMSCPAMAQLLSNTLYYKRFFPYYSFNVLGGLDSEGKGCVF 128
L + L RH YQ + + M A L+S+ Y KRF PY+ V+ GL + K +
Sbjct: 281 TLYQRLVFRHKLYQLREERDMKPETFASLVSSMQYEKRFGPYFCQPVIAGLGDDDKPFIC 460
Query: 129 TYDAVGSYE 137
T DA+G+ E
Sbjct: 461 TMDAIGAKE 487
>TC86040 homologue to GP|17473525|gb|AAL38246.1 proteasome subunit
{Arabidopsis thaliana}, complete
Length = 1151
Score = 75.1 bits (183), Expect = 2e-14
Identities = 41/127 (32%), Positives = 64/127 (50%)
Frame = +2
Query: 11 YDNNGGSCVAIAGSDYCVIAADTRMSTGYNILTRDYSKISHLAEKCVMASSGFQADVKAL 70
+ +NG + VA+ G + IA+D R+ + D+ +IS + +K + SG D + L
Sbjct: 53 FKHNGSAVVAMVGKNCFAIASDRRLGVQLQTIATDFQRISKIHDKLYIGLSGLATDSQTL 232
Query: 71 QKVLSARHLTYQHQHNKQMSCPAMAQLLSNTLYYKRFFPYYSFNVLGGLDSEGKGCVFTY 130
+ L RH YQ + M A L+S Y KRF PY+ V+ GL + K + T
Sbjct: 233 YQRLVFRHKLYQLREELDMKPETFASLVSAMQYEKRFGPYFCQPVIAGLGDDDKPFICTM 412
Query: 131 DAVGSYE 137
DA+G+ E
Sbjct: 413 DAIGAKE 433
>TC86416 homologue to PIR|T04497|T04497 multicatalytic endopeptidase complex
(EC 3.4.99.46) chain PBA1 [imported] - Arabidopsis
thaliana, partial (93%)
Length = 987
Score = 70.9 bits (172), Expect = 4e-13
Identities = 57/209 (27%), Positives = 94/209 (44%), Gaps = 6/209 (2%)
Frame = +3
Query: 15 GGSCVAIAGSDYCVIAADTRMSTGYNILTRDYSKISHLAEKCVMASSGFQADVKALQKVL 74
G + + + + V+ AD+R STG + R KI+ L + + SG AD +V+
Sbjct: 99 GTTIIGVTYNGGVVLGADSRTSTGVYVANRASDKITQLTDNVYVCRSGSAAD----SQVV 266
Query: 75 SARHLTYQHQHNKQMSCPAMAQLLSNTLYYKRFFPYYSFN------VLGGLDSEGKGCVF 128
S + HQH Q+ PA ++ +N + R Y + N ++GG D G ++
Sbjct: 267 SDYVRYFLHQHTIQLGQPATVKVAANLV---RLLAYNNKNNLQTGLIVGGWDKYEGGQIY 437
Query: 129 TYDAVGSYERVGYSSQGSGSTLIMPFLDNQLKSPSPLLLPAQDAVTPLSEAEAVDLVKTV 188
G+ + ++ GSGS+ + F D K +++ EA DLVK
Sbjct: 438 GVPLGGTIVQQPFAIGGSGSSYLYGFFDQAWKD-------------GMTKDEAEDLVKKA 578
Query: 189 FASATERDIYTGDKVEIVILNAGGIHREF 217
+ A RD +G V VI+N+ G+ R F
Sbjct: 579 VSLAIARDGASGGVVRTVIINSEGVTRNF 665
>BI273274 similar to GP|14594933|emb putative beta6 proteasome subunit
{Nicotiana tabacum}, partial (17%)
Length = 649
Score = 65.9 bits (159), Expect = 1e-11
Identities = 31/34 (91%), Positives = 33/34 (96%)
Frame = +3
Query: 58 MASSGFQADVKALQKVLSARHLTYQHQHNKQMSC 91
MASSGFQADVKALQ VLSARHLTYQ+QHNKQ+SC
Sbjct: 471 MASSGFQADVKALQXVLSARHLTYQNQHNKQISC 572
>TC86274 similar to PIR|T51986|T51986 multicatalytic endopeptidase complex
(EC 3.4.99.46) chain PBG1 [imported] - Arabidopsis
thaliana, partial (92%)
Length = 1020
Score = 37.0 bits (84), Expect = 0.006
Identities = 36/161 (22%), Positives = 70/161 (43%), Gaps = 3/161 (1%)
Frame = +2
Query: 1 MTKQHANWSPYDNNGGSCVAIAGSDYCVIAADTRMSTGYNILTRDYSKISHLAEKCVMAS 60
+T+ PY G S VAI D ++AAD S G + + ++ + + ++ +
Sbjct: 131 VTESQRTLYPYVT-GSSVVAIKYKDGILMAADMGGSYGSTLRYKSVERLKPIGKHSLLGA 307
Query: 61 SGFQADVKALQKVLSARHL-TYQHQHNKQMSCPAMAQLLSNTLYYKR--FFPYYSFNVLG 117
SG +D + + + L L + + L+ +Y +R F P ++ VLG
Sbjct: 308 SGEISDFQEIMRYLDELILIDNMWDDGNSLGPKEVHNYLTRVMYNRRNKFNPLWNSLVLG 487
Query: 118 GLDSEGKGCVFTYDAVGSYERVGYSSQGSGSTLIMPFLDNQ 158
G+ +G+ + T +G + + G G+ L P L ++
Sbjct: 488 GV-KKGQKYLGTVSMIGVNYEDNHVATGLGNHLARPILRDE 607
>TC77719 similar to GP|21593260|gb|AAM65209.1 unknown {Arabidopsis
thaliana}, partial (71%)
Length = 1032
Score = 32.0 bits (71), Expect = 0.19
Identities = 15/46 (32%), Positives = 25/46 (53%)
Frame = -1
Query: 41 ILTRDYSKISHLAEKCVMASSGFQADVKALQKVLSARHLTYQHQHN 86
+L R K+SHL C + S +K+L++ +S TY HQ++
Sbjct: 507 VLYRSAPKVSHLVLFCAIVCSHNSTSLKSLRQRVSELQTTYSHQNS 370
>BQ137316
Length = 1059
Score = 32.0 bits (71), Expect = 0.19
Identities = 20/60 (33%), Positives = 27/60 (44%)
Frame = +3
Query: 44 RDYSKISHLAEKCVMASSGFQADVKALQKVLSARHLTYQHQHNKQMSCPAMAQLLSNTLY 103
R +S I V SS AL L +R++T+ H HN+ SCP L +T Y
Sbjct: 507 RVFSTIRLSTLPLVATSSILVQSPPALHLSLCSRYMTHHHTHNRVCSCPHTRCYLHSTPY 686
>TC90328 similar to PIR|B96706|B96706 unknown protein 56300-57889
[imported] - Arabidopsis thaliana, partial (74%)
Length = 877
Score = 29.3 bits (64), Expect = 1.3
Identities = 14/35 (40%), Positives = 16/35 (45%)
Frame = +1
Query: 78 HLTYQHQHNKQMSCPAMAQLLSNTLYYKRFFPYYS 112
H +Q Q NKQ C + L RF PYYS
Sbjct: 136 HSKFQPQRNKQKCCTHFHFVAPTMLLPHRFLPYYS 240
>AJ500104 weakly similar to PIR|T38282|S624 probable proteasome component
precursor - fission yeast (Schizosaccharomyces pombe),
partial (33%)
Length = 424
Score = 27.7 bits (60), Expect = 3.6
Identities = 17/68 (25%), Positives = 28/68 (41%)
Frame = -3
Query: 141 YSSQGSGSTLIMPFLDNQLKSPSPLLLPAQDAVTPLSEAEAVDLVKTVFASATERDIYTG 200
+ + GSGS MP + + L EA+DLVK + D+ +G
Sbjct: 422 FMTMGSGSLAAMPVFERSWRKD-------------LERQEAIDLVKDAIDAGIFNDLGSG 282
Query: 201 DKVEIVIL 208
V+I ++
Sbjct: 281 SNVDINVI 258
>TC77653 similar to GP|20334820|gb|AAM16166.1 At1g12050/F12F1_8 {Arabidopsis
thaliana}, partial (36%)
Length = 778
Score = 27.7 bits (60), Expect = 3.6
Identities = 21/70 (30%), Positives = 32/70 (45%), Gaps = 3/70 (4%)
Frame = -1
Query: 111 YSFNVLGGLDSEGKGCVFT---YDAVGSYERVGYSSQGSGSTLIMPFLDNQLKSPSPLLL 167
YSFN L + + F + +G R Y QG+G T++ + N + PSPL
Sbjct: 574 YSFNTLKKKKKKKRFYSFNNLHHTTIGK-GRGKYFPQGAGGTILPVHVPNPIV*PSPLEY 398
Query: 168 PAQDAVTPLS 177
P + +P S
Sbjct: 397 PVKVTSSPSS 368
>TC77652 similar to GP|20334820|gb|AAM16166.1 At1g12050/F12F1_8 {Arabidopsis
thaliana}, partial (97%)
Length = 1524
Score = 27.7 bits (60), Expect = 3.6
Identities = 21/70 (30%), Positives = 32/70 (45%), Gaps = 3/70 (4%)
Frame = -3
Query: 111 YSFNVLGGLDSEGKGCVFT---YDAVGSYERVGYSSQGSGSTLIMPFLDNQLKSPSPLLL 167
YSFN L + + F + +G R Y QG+G T++ + N + PSPL
Sbjct: 1504 YSFNTLKKKKKKKRFYSFNNLHHTTIGK-GRGKYFPQGAGGTILPVHVPNPIV*PSPLEY 1328
Query: 168 PAQDAVTPLS 177
P + +P S
Sbjct: 1327 PVKVTSSPSS 1298
>TC86021 homologue to GP|19423939|gb|AAL87312.1 putative RNA helicase
{Arabidopsis thaliana}, partial (26%)
Length = 1023
Score = 26.9 bits (58), Expect = 6.2
Identities = 11/25 (44%), Positives = 16/25 (64%)
Frame = +2
Query: 93 AMAQLLSNTLYYKRFFPYYSFNVLG 117
A A ++ + YY RFFP+ S + LG
Sbjct: 764 AFAYVVVSDCYYSRFFPFSSVSCLG 838
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.318 0.133 0.389
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 6,562,867
Number of Sequences: 36976
Number of extensions: 87127
Number of successful extensions: 488
Number of sequences better than 10.0: 34
Number of HSP's better than 10.0 without gapping: 484
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 487
length of query: 223
length of database: 9,014,727
effective HSP length: 93
effective length of query: 130
effective length of database: 5,575,959
effective search space: 724874670
effective search space used: 724874670
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 56 (26.2 bits)
Medicago: description of AC146330.2


