

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC144645.7 - phase: 0
(358 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
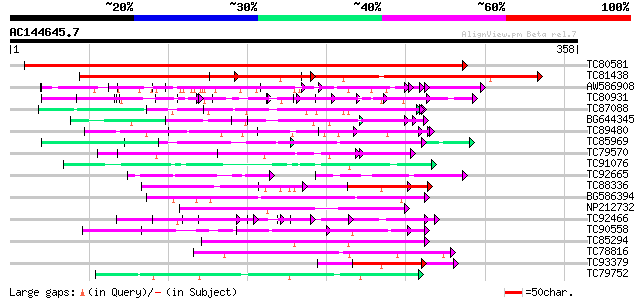
Score E
Sequences producing significant alignments: (bits) Value
TC80581 similar to GP|19683021|gb|AAL92644.1 hypothetical protei... 532 e-152
TC81438 similar to GP|20259253|gb|AAM14362.1 unknown protein {Ar... 144 5e-35
AW586908 weakly similar to GP|2104814|emb| pleiotropic effects o... 89 3e-18
TC80931 similar to GP|14190783|gb|AAF65166.2 putative phloem tra... 71 6e-13
TC87088 similar to PIR|F86399|F86399 protein F17L21.22 [imported... 64 1e-10
BG644345 weakly similar to GP|19697333|gb putative protein poten... 62 3e-10
TC89480 similar to GP|20260684|gb|AAM13240.1 unknown protein {Ar... 60 1e-09
TC85969 similar to GP|21553908|gb|AAM62991.1 unknown {Arabidopsi... 60 2e-09
TC79570 similar to PIR|F86427|F86427 auxin response factor 6 (AR... 58 6e-09
TC91076 similar to PIR|H96747|H96747 unknown protein T10D10.14 [... 57 1e-08
TC92665 homologue to GP|15291583|gb|AAK93060.1 GH28569p {Drosoph... 40 3e-07
TC88336 similar to PIR|T51477|T51477 glutamine-rich protein - Ar... 51 8e-07
BG586394 similar to GP|7295463|gb| CG4835 gene product {Drosophi... 51 8e-07
NP212732 NP212732|AF106929.1|AAD39890.1 putative cell wall protein 50 1e-06
TC92466 similar to GP|11141605|gb|AAG32022.1 LEUNIG {Arabidopsis... 49 2e-06
TC90558 similar to GP|9758584|dbj|BAB09197.1 gb|AAC80623.1~gene_... 49 2e-06
TC85294 similar to GP|2961298|emb|CAA12357.1 hypothetical protei... 49 2e-06
TC78816 similar to GP|14190377|gb|AAK55669.1 AT4g00850/A_TM018A1... 49 3e-06
TC93379 similar to GP|19569994|gb|AAL92295.1 hypothetical protei... 48 5e-06
TC79752 similar to GP|11994670|dbj|BAB02898. gene_id:K7P8.17~unk... 46 2e-05
>TC80581 similar to GP|19683021|gb|AAL92644.1 hypothetical protein
{Dictyostelium discoideum}, partial (5%)
Length = 850
Score = 532 bits (1370), Expect = e-152
Identities = 280/280 (100%), Positives = 280/280 (100%)
Frame = +1
Query: 10 LMTTNGALYAQQLQLSQQAPFNNQQLVGLPQSGQPAMIQNQLLKQIPAVSGSASLLPLQQ 69
LMTTNGALYAQQLQLSQQAPFNNQQLVGLPQSGQPAMIQNQLLKQIPAVSGSASLLPLQQ
Sbjct: 10 LMTTNGALYAQQLQLSQQAPFNNQQLVGLPQSGQPAMIQNQLLKQIPAVSGSASLLPLQQ 189
Query: 70 PQSQQQLASSAQLQQSSLTLNQQQLPQLMQQPKPMGQPQLQQQLQHQQLLQQQLQLQQQY 129
PQSQQQLASSAQLQQSSLTLNQQQLPQLMQQPKPMGQPQLQQQLQHQQLLQQQLQLQQQY
Sbjct: 190 PQSQQQLASSAQLQQSSLTLNQQQLPQLMQQPKPMGQPQLQQQLQHQQLLQQQLQLQQQY 369
Query: 130 QRLQQQLASSAQLQQNSLTLNQQQLPLLMQQPKSMGQPLLQQQQQQLLQQQLQLQQQYQR 189
QRLQQQLASSAQLQQNSLTLNQQQLPLLMQQPKSMGQPLLQQQQQQLLQQQLQLQQQYQR
Sbjct: 370 QRLQQQLASSAQLQQNSLTLNQQQLPLLMQQPKSMGQPLLQQQQQQLLQQQLQLQQQYQR 549
Query: 190 LQQQLASSAQLQQNSSTLNQLPQLMQQQKSMGQPLLQQQLQQQQLLLQQQQLVLQQQQQQ 249
LQQQLASSAQLQQNSSTLNQLPQLMQQQKSMGQPLLQQQLQQQQLLLQQQQLVLQQQQQQ
Sbjct: 550 LQQQLASSAQLQQNSSTLNQLPQLMQQQKSMGQPLLQQQLQQQQLLLQQQQLVLQQQQQQ 729
Query: 250 QQQQRTQLLQQQMQALRIPGPAGHKSLSLTGSQPEATAFG 289
QQQQRTQLLQQQMQALRIPGPAGHKSLSLTGSQPEATAFG
Sbjct: 730 QQQQRTQLLQQQMQALRIPGPAGHKSLSLTGSQPEATAFG 849
>TC81438 similar to GP|20259253|gb|AAM14362.1 unknown protein {Arabidopsis
thaliana}, partial (21%)
Length = 1334
Score = 144 bits (363), Expect = 5e-35
Identities = 87/156 (55%), Positives = 102/156 (64%), Gaps = 4/156 (2%)
Frame = +2
Query: 185 QQYQRLQQQLASSAQLQQNSSTLNQ--LPQLMQQQKSMGQPLLQQQLQQQQLLLQQQQLV 242
QQ+QR QQQLASS QLQQNS TLNQ L Q MQQQKSMGQP L QQ Q QQQQ
Sbjct: 80 QQHQRQQQQLASSGQLQQNSMTLNQQQLSQFMQQQKSMGQPQLHQQQPSPQ---QQQQQQ 250
Query: 243 LQQQQQQQQQQRTQLLQQQMQALRIPGPAGHKSLSLTGSQPEATAFGMTTPGGSLSQGTR 302
L Q QQQ Q Q + QQ + + R+ GP G KS+SLTGSQP+ATA G TTPGGS SQGT
Sbjct: 251 LLQPQQQSQLQASVHQQQHLHSPRVAGPTGQKSISLTGSQPDATASGATTPGGSSSQGTE 430
Query: 303 --TNAMFGQTEIMSVVNELKETRKEVKAIIKILLRF 336
TN + G+ +I +V ++ K +I +LL F
Sbjct: 431 AATNQVLGKRKIQDLVAQVDPQGKLDPEVIDLLLEF 538
Score = 99.4 bits (246), Expect = 2e-21
Identities = 61/102 (59%), Positives = 70/102 (67%), Gaps = 1/102 (0%)
Frame = +2
Query: 45 AMIQNQLLKQIPAVSGSASLLPLQQPQ-SQQQLASSAQLQQSSLTLNQQQLPQLMQQPKP 103
++ Q+Q LKQ+PA+SG AS L LQQ Q QQQLASS QLQQ+S+TLNQQQL Q MQQ K
Sbjct: 11 SLTQSQWLKQMPAMSGPASPLRLQQHQRQQQQLASSGQLQQNSMTLNQQQLSQFMQQQKS 190
Query: 104 MGQPQLQQQLQHQQLLQQQLQLQQQYQRLQQQLASSAQLQQN 145
MGQPQL QQ Q QQQ LQ Q Q QL +S QQ+
Sbjct: 191 MGQPQLHQQQPSPQQQQQQQLLQPQQ---QSQLQASVHQQQH 307
Score = 79.3 bits (194), Expect = 2e-15
Identities = 50/75 (66%), Positives = 52/75 (68%), Gaps = 8/75 (10%)
Frame = +2
Query: 127 QQYQRLQQQLASSAQLQQNSLTLNQQQLPLLMQQPKSMGQPLL-------Q-QQQQQLLQ 178
QQ+QR QQQLASS QLQQNS+TLNQQQL MQQ KSMGQP L Q QQQQQLLQ
Sbjct: 80 QQHQRQQQQLASSGQLQQNSMTLNQQQLSQFMQQQKSMGQPQLHQQQPSPQQQQQQQLLQ 259
Query: 179 QQLQLQQQYQRLQQQ 193
Q Q Q Q QQQ
Sbjct: 260 PQQQSQLQASVHQQQ 304
>AW586908 weakly similar to GP|2104814|emb| pleiotropic effects on cellular
differentiation and slug behaviour {Dictyostelium
discoideum}, partial (8%)
Length = 668
Score = 88.6 bits (218), Expect = 3e-18
Identities = 88/214 (41%), Positives = 103/214 (48%), Gaps = 21/214 (9%)
Frame = +2
Query: 63 SLLPL------QQPQSQQQLASSAQLQQSSLTLNQQQLPQLMQQPKPMGQPQLQQQLQH- 115
SLLP+ QQ Q QQQLA S L Q QQQ PQL QQ QPQ QQQ Q
Sbjct: 56 SLLPMPLDQLQQQQQRQQQLAGSQNLPQP-----QQQQPQLTQQSPQ--QPQQQQQHQQP 214
Query: 116 -------------QQLLQQQLQLQQQYQRLQQQLAS-SAQLQQNSLTLNQQQLPLLMQQP 161
Q+ Q +Q Y +LQQQL S S Q QQN + + L +
Sbjct: 215 CQNTTMNNGTIGSNQIPNQCVQQPVTYSQLQQQLLSGSMQSQQNLQSAGKNGLMMTSLPQ 394
Query: 162 KSMGQPLLQQQQQQLLQQQLQLQQQYQRLQQQLASSAQLQQNSSTLNQLPQLMQQQKSMG 221
S Q + QQQ LLQ+Q QQ +LQQ SS QL Q S L + PQ Q +++
Sbjct: 395 DSQFQQQIDQQQAGLLQRQ----QQQNQLQQ---SSLQLLQQSM-LQRGPQQPQMSQTVP 550
Query: 222 QPLLQQQLQQQQLLLQQQQLVLQQQQQQQQQQRT 255
Q + QQ Q Q L QQQ QQQQQQQQ T
Sbjct: 551 QNISDQQQQLQLLQKLQQQ---QQQQQQQQPLST 643
Score = 81.6 bits (200), Expect = 4e-16
Identities = 75/181 (41%), Positives = 84/181 (45%), Gaps = 17/181 (9%)
Frame = +2
Query: 99 QQPKPMGQPQLQQQLQHQQLLQQQLQLQQQYQRLQQQLASSAQLQQNSLTLNQQQLPLLM 158
QQP Q QQQ + Q LL L QQ Q+ QQQLA S L Q QQQ P L
Sbjct: 2 QQPPASWSQQQQQQQKMQSLLPMPLDQLQQQQQRQQQLAGSQNLPQP-----QQQQPQLT 166
Query: 159 QQPKSMGQPLLQQQQQQLLQ----------------QQLQLQQQYQRLQQQLAS-SAQLQ 201
QQ S QP QQQ QQ Q Q +Q Y +LQQQL S S Q Q
Sbjct: 167 QQ--SPQQPQQQQQHQQPCQNTTMNNGTIGSNQIPNQCVQQPVTYSQLQQQLLSGSMQSQ 340
Query: 202 QNSSTLNQLPQLMQQQKSMGQPLLQQQLQQQQLLLQQQQLVLQQQQQQQQQQRTQLLQQQ 261
QN + + +M Q QQQ+ QQQ L Q+ QQQQ Q QQ QLLQQ
Sbjct: 341 QNLQSAGKNGLMMTSLPQDSQ--FQQQIDQQQAGLLQR----QQQQNQLQQSSLQLLQQS 502
Query: 262 M 262
M
Sbjct: 503 M 505
Score = 78.6 bits (192), Expect = 4e-15
Identities = 85/214 (39%), Positives = 100/214 (46%), Gaps = 30/214 (14%)
Frame = +2
Query: 69 QPQSQQQLASSA------QLQQSSLTLNQ----QQLPQLMQQPKPMGQ---PQLQQQLQH 115
Q Q QQQ S QLQQ Q Q LPQ QQ + Q Q QQQ QH
Sbjct: 26 QQQQQQQKMQSLLPMPLDQLQQQQQRQQQLAGSQNLPQPQQQQPQLTQQSPQQPQQQQQH 205
Query: 116 QQLLQ----------------QQLQLQQQYQRLQQQLAS-SAQLQQNSLTLNQQQLPLLM 158
QQ Q Q +Q Y +LQQQL S S Q QQN + + L +
Sbjct: 206 QQPCQNTTMNNGTIGSNQIPNQCVQQPVTYSQLQQQLLSGSMQSQQNLQSAGKNGLMMTS 385
Query: 159 QQPKSMGQPLLQQQQQQLLQQQLQLQQQYQRLQQQLASSAQLQQNSSTLNQLPQLMQQQK 218
S Q + QQQ LLQ+ QQQ +LQQ SS QL Q S L + PQ Q +
Sbjct: 386 LPQDSQFQQQIDQQQAGLLQR----QQQQNQLQQ---SSLQLLQ-QSMLQRGPQQPQMSQ 541
Query: 219 SMGQPLLQQQLQQQQLLLQQQQLVLQQQQQQQQQ 252
++ Q + QQ QQ QLL + QQ QQQQQQQQQ
Sbjct: 542 TVPQNISDQQ-QQLQLLQKLQQ---QQQQQQQQQ 631
Score = 78.2 bits (191), Expect = 5e-15
Identities = 76/206 (36%), Positives = 96/206 (45%), Gaps = 28/206 (13%)
Frame = +2
Query: 21 QLQLSQQAPFNNQQLVGLPQSGQPAMIQNQLLKQIPAVSGSASLLPLQQPQSQQQLASSA 80
QLQ QQ QQL G QP Q QL +Q P QQPQ QQQ
Sbjct: 80 QLQQQQQ---RQQQLAGSQNLPQPQQQQPQLTQQSP-----------QQPQQQQQHQQPC 217
Query: 81 Q-LQQSSLTLNQQQLP-QLMQQPKPMGQPQLQQQL-----QHQQLLQQ------------ 121
Q ++ T+ Q+P Q +QQP + QLQQQL Q QQ LQ
Sbjct: 218 QNTTMNNGTIGSNQIPNQCVQQP--VTYSQLQQQLLSGSMQSQQNLQSAGKNGLMMTSLP 391
Query: 122 -----QLQLQQQYQRLQQQLASSAQLQQNSLTLNQQQL----PLLMQQPKSMGQPLLQQQ 172
Q Q+ QQ L Q+ QLQQ+SL L QQ + P Q +++ Q + QQ
Sbjct: 392 QDSQFQQQIDQQQAGLLQRQQQQNQLQQSSLQLLQQSMLQRGPQQPQMSQTVPQNISDQQ 571
Query: 173 QQQLLQQQLQLQQQYQRLQQQLASSA 198
QQ L Q+LQ QQQ Q+ QQ L++S+
Sbjct: 572 QQLQLLQKLQQQQQQQQQQQPLSTSS 649
Score = 73.2 bits (178), Expect = 1e-13
Identities = 80/215 (37%), Positives = 95/215 (43%), Gaps = 40/215 (18%)
Frame = +2
Query: 91 QQQLPQLMQQPKPMGQPQLQQQLQHQQLL---QQQLQLQQQYQRLQQQLASSAQLQQ--- 144
QQQ Q MQ PM QLQQQ Q QQ L Q Q QQQ +L QQ Q QQ
Sbjct: 29 QQQQQQKMQSLLPMPLDQLQQQQQRQQQLAGSQNLPQPQQQQPQLTQQSPQQPQQQQQHQ 208
Query: 145 --------NSLTLNQQQLP-LLMQQPKSMGQPLLQQQQQQLLQQQLQLQQQYQRLQQQLA 195
N+ T+ Q+P +QQP + Q QQQLL +Q QQ Q +
Sbjct: 209 QPCQNTTMNNGTIGSNQIPNQCVQQPVTYSQ-----LQQQLLSGSMQSQQNLQSAGKNGL 373
Query: 196 SSAQLQQNSSTLNQLPQ----LMQQQKSMGQPLLQQ---QLQQQQLLL------------ 236
L Q+S Q+ Q L+Q+Q+ Q LQQ QL QQ +L
Sbjct: 374 MMTSLPQDSQFQQQIDQQQAGLLQRQQQ--QNQLQQSSLQLLQQSMLQRGPQQPQMSQTV 547
Query: 237 ------QQQQLVLQQQQQQQQQQRTQLLQQQMQAL 265
QQQQL L Q+ QQQQQQ QQQ Q L
Sbjct: 548 PQNISDQQQQLQLLQKLQQQQQQ-----QQQQQPL 637
Score = 63.2 bits (152), Expect = 2e-10
Identities = 67/181 (37%), Positives = 81/181 (44%), Gaps = 13/181 (7%)
Frame = +2
Query: 20 QQLQLSQQAPFNNQQLVGLPQSGQPAMIQNQLL--KQIPAVSGSASLLPLQQPQSQQQLA 77
QQ QL+QQ+P QQ Q Q + N + QIP P+ Q QQQL
Sbjct: 149 QQPQLTQQSPQQPQQQQQHQQPCQNTTMNNGTIGSNQIP---NQCVQQPVTYSQLQQQLL 319
Query: 78 SSAQLQQSSLT------LNQQQLPQLMQQPKPMGQPQ---LQQQLQHQQLLQQQLQLQQQ 128
S + Q +L L LPQ Q + + Q Q LQ+Q Q QL Q LQL QQ
Sbjct: 320 SGSMQSQQNLQSAGKNGLMMTSLPQDSQFQQQIDQQQAGLLQRQQQQNQLQQSSLQLLQQ 499
Query: 129 --YQRLQQQLASSAQLQQNSLTLNQQQLPLLMQQPKSMGQPLLQQQQQQLLQQQLQLQQQ 186
QR QQ S + QN ++ QQQL LL Q L QQQQQQ QQ L
Sbjct: 500 SMLQRGPQQPQMSQTVPQN-ISDQQQQLQLL--------QKLQQQQQQQQQQQPLSTSSH 652
Query: 187 Y 187
+
Sbjct: 653 F 655
Score = 47.0 bits (110), Expect = 1e-05
Identities = 49/147 (33%), Positives = 61/147 (41%), Gaps = 5/147 (3%)
Frame = +2
Query: 159 QQPKSMGQPLLQQQQQQLLQQQL-----QLQQQYQRLQQQLASSAQLQQNSSTLNQLPQL 213
Q P S Q QQQQQQ +Q L QLQQQ QR QQQLA S
Sbjct: 5 QPPASWSQ---QQQQQQKMQSLLPMPLDQLQQQQQR-QQQLAGS---------------- 124
Query: 214 MQQQKSMGQPLLQQQLQQQQLLLQQQQLVLQQQQQQQQQQRTQLLQQQMQALRIPGPAGH 273
Q L Q Q QQ QL Q Q QQQQ QQ Q T + + + +IP
Sbjct: 125 --------QNLPQPQQQQPQLTQQSPQQPQQQQQHQQPCQNTTMNNGTIGSNQIPNQCVQ 280
Query: 274 KSLSLTGSQPEATAFGMTTPGGSLSQG 300
+ ++ + Q + + M + S G
Sbjct: 281 QPVTYSQLQQQLLSGSMQSQQNLQSAG 361
Score = 30.8 bits (68), Expect = 0.84
Identities = 20/76 (26%), Positives = 30/76 (39%)
Frame = +2
Query: 18 YAQQLQLSQQAPFNNQQLVGLPQSGQPAMIQNQLLKQIPAVSGSASLLPLQQPQSQQQLA 77
+ QQ+ Q QQ Q ++Q +L++ P + +P QQQL
Sbjct: 404 FQQQIDQQQAGLLQRQQQQNQLQQSSLQLLQQSMLQRGPQQPQMSQTVPQNISDQQQQLQ 583
Query: 78 SSAQLQQSSLTLNQQQ 93
+LQQ QQQ
Sbjct: 584 LLQKLQQQQQQQQQQQ 631
>TC80931 similar to GP|14190783|gb|AAF65166.2 putative phloem transcription
factor M1 {Apium graveolens}, partial (67%)
Length = 1193
Score = 71.2 bits (173), Expect = 6e-13
Identities = 52/117 (44%), Positives = 64/117 (54%), Gaps = 1/117 (0%)
Frame = +2
Query: 180 QLQLQQQYQRLQQQLASSAQLQQNSSTLNQLPQLMQQQKSMGQPLLQQQLQQQQLLLQQQ 239
QLQ Q Q Q+ QQQL Q QQ S Q+PQL QQQ+ + P LQQQ Q QL QQQ
Sbjct: 563 QLQQQMQQQQQQQQLPQLQQNQQLSQIQQQIPQLQQQQQQL--PQLQQQ-QLSQLQQQQQ 733
Query: 240 QL-VLQQQQQQQQQQRTQLLQQQMQALRIPGPAGHKSLSLTGSQPEATAFGMTTPGG 295
QL LQQ Q QQ Q+ Q++ M + GP G + SQ + ++ G T GG
Sbjct: 734 QLPQLQQLQHQQLPQQQQMVGAGMGQTYVQGPGGRSQMV---SQGQVSSQGATNIGG 895
Score = 66.6 bits (161), Expect = 1e-11
Identities = 57/118 (48%), Positives = 63/118 (53%), Gaps = 2/118 (1%)
Frame = +2
Query: 107 PQLQQQLQHQQLLQQ--QLQLQQQYQRLQQQLASSAQLQQNSLTLNQQQLPLLMQQPKSM 164
PQLQQQ+Q QQ QQ QLQ QQ ++QQQ+ QLQQ QQQLP L QQ S
Sbjct: 560 PQLQQQMQQQQQQQQLPQLQQNQQLSQIQQQIP---QLQQQ-----QQQLPQLQQQQLSQ 715
Query: 165 GQPLLQQQQQQLLQQQLQLQQQYQRLQQQLASSAQLQQNSSTLNQLPQLMQQQKSMGQ 222
LQQQQQQL Q Q QLQ Q QQQ+ + Q T Q P Q S GQ
Sbjct: 716 ----LQQQQQQLPQLQ-QLQHQQLPQQQQMVGAGMGQ----TYVQGPGGRSQMVSQGQ 862
Score = 57.0 bits (136), Expect = 1e-08
Identities = 45/100 (45%), Positives = 52/100 (52%)
Frame = +2
Query: 67 LQQPQSQQQLASSAQLQQSSLTLNQQQLPQLMQQPKPMGQPQLQQQLQHQQLLQQQLQLQ 126
+QQ Q QQQL Q QQ L+ QQQ+PQL QQ + + PQLQQQ Q QL QQQ QL
Sbjct: 578 MQQQQQQQQLPQLQQNQQ--LSQIQQQIPQLQQQQQQL--PQLQQQ-QLSQLQQQQQQLP 742
Query: 127 QQYQRLQQQLASSAQLQQNSLTLNQQQLPLLMQQPKSMGQ 166
Q Q QQL Q+ + Q P Q S GQ
Sbjct: 743 QLQQLQHQQLPQQQQMVGAGMGQTYVQGPGGRSQMVSQGQ 862
Score = 55.8 bits (133), Expect = 2e-08
Identities = 45/112 (40%), Positives = 54/112 (48%), Gaps = 1/112 (0%)
Frame = +2
Query: 129 YQRLQQQLASSAQLQQNSLTLNQQQLPLLMQQPKSMGQPLLQQQQQQLLQQQLQLQQQYQ 188
+ +LQQQ+ Q QQ QQL + QQ P LQQQQQQL Q Q Q Q Q
Sbjct: 557 FPQLQQQMQQQQQQQQLPQLQQNQQLSQIQQQ-----IPQLQQQQQQLPQLQQQQLSQLQ 721
Query: 189 RLQQQLASSAQLQQNSSTLNQLPQLMQQ-QKSMGQPLLQQQLQQQQLLLQQQ 239
+ QQQL QLQ QLPQ Q MGQ +Q + Q++ Q Q
Sbjct: 722 QQQQQLPQLQQLQH-----QQLPQQQQMVGAGMGQTYVQGPGGRSQMVSQGQ 862
Score = 53.9 bits (128), Expect = 9e-08
Identities = 50/119 (42%), Positives = 57/119 (47%), Gaps = 4/119 (3%)
Frame = +2
Query: 70 PQSQQQLASSAQLQQSSLTLNQQQLPQLMQQPKPMGQPQLQQQLQHQQLLQQQLQLQQQY 129
PQ QQQ+ Q QQ QQL Q+ QQ PQLQQQ QQL Q Q QQQ
Sbjct: 560 PQLQQQMQQQQQQQQLPQLQQNQQLSQIQQQI-----PQLQQQ--QQQLPQLQ---QQQL 709
Query: 130 QRLQQQLASSAQLQQNSLTLNQQQLPLLMQQ-PKSMGQPLLQ---QQQQQLLQQQLQLQ 184
+LQQQ QLQQ L QQLP Q MGQ +Q + Q + Q Q+ Q
Sbjct: 710 SQLQQQQQQLPQLQQ----LQHQQLPQQQQMVGAGMGQTYVQGPGGRSQMVSQGQVSSQ 874
Score = 52.0 bits (123), Expect = 4e-07
Identities = 46/116 (39%), Positives = 56/116 (47%), Gaps = 2/116 (1%)
Frame = +2
Query: 93 QLPQLMQQPKPMGQ-PQLQQQLQHQQLLQQQLQLQQQYQRLQQ-QLASSAQLQQNSLTLN 150
QL Q MQQ + Q PQLQQ Q Q+ QQ QLQQQ Q+L Q Q +QLQQ
Sbjct: 563 QLQQQMQQQQQQQQLPQLQQNQQLSQIQQQIPQLQQQQQQLPQLQQQQLSQLQQQQ---- 730
Query: 151 QQQLPLLMQQPKSMGQPLLQQQQQQLLQQQLQLQQQYQRLQQQLASSAQLQQNSST 206
QQLP L Q + L QQQQ + Q Q + Q+ S Q+ +T
Sbjct: 731 -QQLPQLQQ----LQHQQLPQQQQMVGAGMGQTYVQGPGGRSQMVSQGQVSSQGAT 883
Score = 51.2 bits (121), Expect = 6e-07
Identities = 39/82 (47%), Positives = 46/82 (55%), Gaps = 2/82 (2%)
Frame = +2
Query: 43 QPAMIQNQLLKQIPAVSGSASLLPLQQ--PQSQQQLASSAQLQQSSLTLNQQQLPQLMQQ 100
Q M Q Q +Q+P + + L +QQ PQ QQQ QLQQ QQL QL QQ
Sbjct: 569 QQQMQQQQQQQQLPQLQQNQQLSQIQQQIPQLQQQQQQLPQLQQ-------QQLSQLQQQ 727
Query: 101 PKPMGQPQLQQQLQHQQLLQQQ 122
+ + PQLQQ LQHQQL QQQ
Sbjct: 728 QQQL--PQLQQ-LQHQQLPQQQ 784
Score = 49.3 bits (116), Expect = 2e-06
Identities = 44/99 (44%), Positives = 50/99 (50%), Gaps = 1/99 (1%)
Frame = +2
Query: 68 QQPQSQQQLASSAQLQQSS-LTLNQQQLPQLMQQPKPMGQPQLQQQLQHQQLLQQQLQLQ 126
QQ Q QQQ QLQQ+ L+ QQQ+PQL QQ + + PQLQQQ QL QLQ Q
Sbjct: 572 QQMQQQQQQQQLPQLQQNQQLSQIQQQIPQLQQQQQQL--PQLQQQ----QL--SQLQQQ 727
Query: 127 QQYQRLQQQLASSAQLQQNSLTLNQQQLPLLMQQPKSMG 165
QQQL QLQ L QQ + M Q G
Sbjct: 728 ------QQQLPQLQQLQHQQLPQQQQMVGAGMGQTYVQG 826
Score = 40.8 bits (94), Expect = 8e-04
Identities = 35/102 (34%), Positives = 42/102 (40%), Gaps = 1/102 (0%)
Frame = +2
Query: 21 QLQLSQQAPFNNQQLVGLPQSGQPAMIQNQLLKQIPAVSGSASLLPLQQPQSQQQLASSA 80
QLQ Q QQL L Q+ Q + IQ +QIP + LP Q Q QL
Sbjct: 563 QLQQQMQQQQQQQQLPQLQQNQQLSQIQ----QQIPQLQQQQQQLPQLQQQQLSQLQQQQ 730
Query: 81 QLQQSSLTLNQQQLPQLMQQ-PKPMGQPQLQQQLQHQQLLQQ 121
Q L QQLPQ Q MGQ +Q Q++ Q
Sbjct: 731 QQLPQLQQLQHQQLPQQQQMVGAGMGQTYVQGPGGRSQMVSQ 856
Score = 40.8 bits (94), Expect = 8e-04
Identities = 33/81 (40%), Positives = 40/81 (48%), Gaps = 8/81 (9%)
Frame = +2
Query: 194 LASSAQLQQNSSTLNQLPQLMQQQKSMGQPLLQQQLQQQQ----LLLQQQQLVLQQQQ-- 247
L SS+ + NS+ N + + Q Q Q QQQQ L Q QQL QQQ
Sbjct: 479 LVSSSSNRCNSNKCNNTSRCNRNSSIFPQLQQQMQQQQQQQQLPQLQQNQQLSQIQQQIP 658
Query: 248 --QQQQQQRTQLLQQQMQALR 266
QQQQQQ QL QQQ+ L+
Sbjct: 659 QLQQQQQQLPQLQQQQLSQLQ 721
Score = 40.4 bits (93), Expect = 0.001
Identities = 41/150 (27%), Positives = 63/150 (41%), Gaps = 5/150 (3%)
Frame = +1
Query: 4 SNLASQLMTTNGALYAQQLQLSQQAPFNNQQLVGLPQSGQPAMIQNQLLKQIPAVSGSAS 63
S+ AS+ + N Q ++L Q NN+Q VG A++
Sbjct: 190 SSSASETLAANWPPVMQIVRLISQDHMNNKQYVGKAD-----------FLVFRAMNPHGF 336
Query: 64 LLPLQQPQSQQQLASSAQLQQSSLTLNQQQ-----LPQLMQQPKPMGQPQLQQQLQHQQL 118
L LQ+ ++L + QL +L L+ + L + +PQL Q Q QQ+
Sbjct: 337 LGQLQE----KKLCAVIQLPSQTLLLSVSDKACRLIGMLFPGDMVVFKPQLSGQQQQQQM 504
Query: 119 LQQQLQLQQQYQRLQQQLASSAQLQQNSLT 148
QQQ+Q QQ Q QQ L++ A + T
Sbjct: 505 QQQQMQQHQQMQSQQQHLSTIAATDATTTT 594
Score = 37.7 bits (86), Expect = 0.007
Identities = 51/199 (25%), Positives = 88/199 (43%), Gaps = 12/199 (6%)
Frame = +1
Query: 20 QQLQLSQQAPFNNQQLVGLPQSGQPAMIQNQLLKQIP-AVSGSASLLPLQQPQSQQQLAS 78
QQ+Q Q NN + + Q A Q++ +K ++SG P+ + + +S
Sbjct: 16 QQVQSGMQPL*NNAAAANMTLTQQTASSQSKYIKVWEGSLSGQRQGQPVFITKLEGYRSS 195
Query: 79 SAQLQQSSLTLNQQQLPQLMQQPKPMGQPQL--QQQLQHQQLLQQQLQLQQQY--QRLQQ 134
SA +L N P +MQ + + Q + +Q + L + + Q ++
Sbjct: 196 SAS---ETLAANW---PPVMQIVRLISQDHMNNKQYVGKADFLVFRAMNPHGFLGQLQEK 357
Query: 135 QLASSAQLQQNSLTLNQQQ-----LPLLMQQPKSMGQPLL--QQQQQQLLQQQLQLQQQY 187
+L + QL +L L+ + +L + +P L QQQQQQ+ QQQ+Q QQ
Sbjct: 358 KLCAVIQLPSQTLLLSVSDKACRLIGMLFPGDMVVFKPQLSGQQQQQQMQQQQMQQHQQM 537
Query: 188 QRLQQQLASSAQLQQNSST 206
Q QQ L++ A ++T
Sbjct: 538 QSQQQHLSTIAATDATTTT 594
Score = 34.3 bits (77), Expect = 0.076
Identities = 28/101 (27%), Positives = 43/101 (41%)
Frame = +1
Query: 222 QPLLQQQLQQQQLLLQQQQLVLQQQQQQQQQQRTQLLQQQMQALRIPGPAGHKSLSLTGS 281
+P L Q QQQQ +QQQQ +QQ QQ Q QQ+ A + + + +
Sbjct: 466 KPQLSGQQQQQQ--MQQQQ--MQQHQQMQSQQQHLSTIAATDATTTTTATASTAATESAA 633
Query: 282 QPEATAFGMTTPGGSLSQGTRTNAMFGQTEIMSVVNELKET 322
P++TA T + + T T A F + + + T
Sbjct: 634 FPDSTADSTTPATAAATPSTSTAAAFSAAATAATASAVTAT 756
>TC87088 similar to PIR|F86399|F86399 protein F17L21.22 [imported] -
Arabidopsis thaliana, partial (4%)
Length = 2997
Score = 63.5 bits (153), Expect = 1e-10
Identities = 57/154 (37%), Positives = 71/154 (46%), Gaps = 14/154 (9%)
Frame = +3
Query: 123 LQLQQ-QYQRLQQQLASSAQLQQNSLTLNQQQLPLLMQQPKSMGQPLLQQQQQQLLQQQL 181
L LQ+ Q +RLQQQ A Q QQ + L QQQ L QQ LQQQQ + LQQQ
Sbjct: 342 LMLQKLQKERLQQQQAERLQ-QQQAERLQQQQAERLQQQQAER----LQQQQAERLQQQT 506
Query: 182 QLQQQYQRLQQ-----QLASSAQLQQNSSTLNQLPQ--------LMQQQKSMGQPLLQQQ 228
+ + + + Q N S + Q+ Q L + Q Q +QQQ
Sbjct: 507 NISSHFPAYLNGSDLDRFPGFSPSQSNKSGIQQMMQNPGTDFERLFELQAQQRQLEIQQQ 686
Query: 229 LQQQQLLLQQQQLVLQQQQQQQQQQRTQLLQQQM 262
Q LLQQ +L QQQ Q QQ QL+ QQM
Sbjct: 687 DMHHQQLLQQLKLQPQQQSQVQQLLLEQLMHQQM 788
Score = 55.5 bits (132), Expect = 3e-08
Identities = 73/257 (28%), Positives = 100/257 (38%), Gaps = 15/257 (5%)
Frame = +3
Query: 19 AQQLQLSQQAPFNNQQLVGLPQSGQPAMIQNQLLKQIPAVSGSASLLPLQQPQSQQQLAS 78
A++LQ Q QQ L Q Q +Q Q +++ + +S P S
Sbjct: 387 AERLQQQQAERLQQQQAERLQQQ-QAERLQQQQAERLQQQTNISSHFPAYLNGSDLDRFP 563
Query: 79 SAQLQQSSLTLNQQQLPQLMQQP-----------KPMGQPQLQQQLQHQQLLQQQLQLQQ 127
QS N+ + Q+MQ P Q ++QQQ H Q L QQL+LQ
Sbjct: 564 GFSPSQS----NKSGIQQMMQNPGTDFERLFELQAQQRQLEIQQQDMHHQQLLQQLKLQP 731
Query: 128 QYQRLQQQLASSAQLQQNSLTLNQQQLPLLMQQPKSMGQPLLQQQQQQLLQQQLQLQQQY 187
Q Q QQL L +Q + M P + GQ + LL Q+QL++
Sbjct: 732 QQQSQVQQL------------LLEQLMHQQMSDP-NFGQSKHDPSRDNLL-DQVQLRRYL 869
Query: 188 QRLQQQLASSAQLQQNSSTLNQ----LPQLMQQQKSMGQPLLQQQLQQQQLLLQQQQLVL 243
LQQ S L + Q L +Q + + LL Q + +L QQL
Sbjct: 870 HDLQQNPHSLGHLDPSMEQFIQANIGLNAAQGRQADLSELLL--QARHGNILPSDQQLRF 1043
Query: 244 QQQQQQQQQQRTQLLQQ 260
QQ Q Q QQ L QQ
Sbjct: 1044QQDQLQAQQLSMALKQQ 1094
Score = 51.6 bits (122), Expect = 5e-07
Identities = 63/203 (31%), Positives = 85/203 (41%), Gaps = 26/203 (12%)
Frame = +3
Query: 87 LTLNQQQLPQLMQQPKPMGQPQLQQQLQHQQLLQQQLQ--LQQQYQRLQQQLASSAQLQQ 144
L L + Q +L QQ Q + QQ Q ++L QQQ + QQQ +RLQQQ A Q Q
Sbjct: 342 LMLQKLQKERLQQQ-----QAERLQQQQAERLQQQQAERLQQQQAERLQQQQAERLQQQT 506
Query: 145 NSLT---------------------LNQQQLPLLMQQPKSMGQPLLQQQQQQLLQQQLQL 183
N + N+ + +MQ P + + L + Q Q Q+QL++
Sbjct: 507 NISSHFPAYLNGSDLDRFPGFSPSQSNKSGIQQMMQNPGTDFERLFELQAQ---QRQLEI 677
Query: 184 QQQ---YQRLQQQLASSAQLQQNSSTLNQLPQLMQQQKSMGQPLLQQQLQQQQLLLQQQQ 240
QQQ +Q+L QQL Q Q L L QLM QQ S + + LL Q Q
Sbjct: 678 QQQDMHHQQLLQQLKLQPQQQSQVQQL-LLEQLMHQQMSDPNFGQSKHDPSRDNLLDQVQ 854
Query: 241 LVLQQQQQQQQQQRTQLLQQQMQ 263
L QQ L M+
Sbjct: 855 LRRYLHDLQQNPHSLGHLDPSME 923
Score = 37.4 bits (85), Expect = 0.009
Identities = 46/155 (29%), Positives = 66/155 (41%), Gaps = 11/155 (7%)
Frame = +3
Query: 118 LLQQQLQLQQQYQRLQQQLASSAQLQQNSLT--LNQQQLPLLMQQPKSMGQPLLQQQQQQ 175
LL +L+ Q +R Q +S Q+ T L + P Q SMG + Q +
Sbjct: 45 LLMSELRDGSQLRRAQSSNSSLRLGDQSHFTDPLIDRDAPFTDQS--SMGGMVNQSSFRD 218
Query: 176 L------LQQQLQLQQQYQRLQQQLASSAQLQQNSSTLNQLPQLMQQQKSMGQPLLQQQL 229
+ + Q+ L+ Q S N+ + L + QK Q ++L
Sbjct: 219 TWTDEYGINRHFNPNQRVGSLEDQFLSRIGPNFNNFDVADHLMLQKLQKERLQQQQAERL 398
Query: 230 QQQQL-LLQQQQLVLQQQQQQQ--QQQRTQLLQQQ 261
QQQQ LQQQQ QQQQ + QQQ+ + LQQQ
Sbjct: 399 QQQQAERLQQQQAERLQQQQAERLQQQQAERLQQQ 503
Score = 37.0 bits (84), Expect = 0.012
Identities = 26/51 (50%), Positives = 33/51 (63%), Gaps = 4/51 (7%)
Frame = +3
Query: 220 MGQPLLQQQLQQQQL--LLQQQQLVLQQQQQQ--QQQQRTQLLQQQMQALR 266
M Q L +++LQQQQ L QQQ LQQQQ + QQQQ +L QQQ + L+
Sbjct: 345 MLQKLQKERLQQQQAERLQQQQAERLQQQQAERLQQQQAERLQQQQAERLQ 497
Score = 33.1 bits (74), Expect = 0.17
Identities = 22/45 (48%), Positives = 31/45 (68%), Gaps = 2/45 (4%)
Frame = +3
Query: 224 LLQQQLQQQQLLLQQQQLVLQQQQQQ--QQQQRTQLLQQQMQALR 266
L+ Q+LQ+++L QQQ LQQQQ + QQQQ +L QQQ + L+
Sbjct: 342 LMLQKLQKERLQ-QQQAERLQQQQAERLQQQQAERLQQQQAERLQ 473
>BG644345 weakly similar to GP|19697333|gb putative protein potential
transcriptional repressor Not4hp - Mus musculus, partial
(7%)
Length = 635
Score = 62.4 bits (150), Expect = 3e-10
Identities = 49/116 (42%), Positives = 57/116 (48%), Gaps = 2/116 (1%)
Frame = +1
Query: 151 QQQLPLLMQQPKSMGQPLLQQQQQQLLQQQLQLQQQYQRLQQQLASSAQL--QQNSSTLN 208
Q Q P L Q P QQQ Q +Q QQ AS Q QQ S +
Sbjct: 139 QAQQPSLFQTPG-------QQQASPFQTPGQQQSSPFQTPGQQQASPFQTPGQQQPSPFS 297
Query: 209 QLPQLMQQQKSMGQPLLQQQLQQQQLLLQQQQLVLQQQQQQQQQQRTQLLQQQMQA 264
Q+ QQQ+ + Q QQQ QQQQL QQQQ + QQQQ Q QQQ+ QLLQQ +
Sbjct: 298 QITPFSQQQQQL-QFQQQQQQQQQQLQQQQQQQLQQQQQLQLQQQQQQLLQQHQSS 462
Score = 61.6 bits (148), Expect = 4e-10
Identities = 64/198 (32%), Positives = 74/198 (37%), Gaps = 12/198 (6%)
Frame = +1
Query: 39 PQSGQPAMIQNQLLKQIPAVSGSASLLPLQQPQSQQQ--LASSAQLQQSSLTLNQQQLPQ 96
PQ+ QP++ Q G P Q P QQ + Q Q S QQ P
Sbjct: 136 PQAQQPSLFQTP---------GQQQASPFQTPGQQQSSPFQTPGQQQASPFQTPGQQQPS 288
Query: 97 LMQQPKPMGQPQLQQQLQHQQLLQQQLQLQQQYQRLQQQLASSAQLQQNSLTLNQQQLPL 156
Q P Q Q Q Q Q QQ QQQ QQQ Q+LQQQ QLQ
Sbjct: 289 PFSQITPFSQQQQQLQFQQQQQQQQQQLQQQQQQQLQQQ----QQLQ------------- 417
Query: 157 LMQQPKSMGQPLLQQQQQQLLQQQLQLQQQYQRL----------QQQLASSAQLQQNSST 206
LQQQQQQLLQQ QQQ L + A L +
Sbjct: 418 ------------LQQQQQQLLQQHQSSQQQQLYLFTNDKTPATYSTKWADLHSLLPHPLL 561
Query: 207 LNQLPQLMQQQKSMGQPL 224
L+ LP + QQ + PL
Sbjct: 562 LHLLPCSLLQQCQLQSPL 615
Score = 55.8 bits (133), Expect = 2e-08
Identities = 48/119 (40%), Positives = 52/119 (43%), Gaps = 2/119 (1%)
Frame = +1
Query: 141 QLQQNSL--TLNQQQLPLLMQQPKSMGQPLLQQQQQQLLQQQLQLQQQYQRLQQQLASSA 198
Q QQ SL T QQQ + P QQQ Q QQQ Q S
Sbjct: 139 QAQQPSLFQTPGQQQASPFQTPGQQQSSPFQTPGQQQASPFQTPGQQQPSPFSQITPFSQ 318
Query: 199 QLQQNSSTLNQLPQLMQQQKSMGQPLLQQQLQQQQLLLQQQQLVLQQQQQQQQQQRTQL 257
Q QQ L Q QQQ+ + Q QQ QQQQL LQQQQ L QQ Q QQQ+ L
Sbjct: 319 QQQQ----LQFQQQQQQQQQQLQQQQQQQLQQQQQLQLQQQQQQLLQQHQSSQQQQLYL 483
Score = 55.5 bits (132), Expect = 3e-08
Identities = 62/154 (40%), Positives = 65/154 (41%)
Frame = +1
Query: 99 QQPKPMGQPQLQQQLQHQQLLQQQLQLQQQYQRLQQQLASSAQLQQNSLTLNQQQLPLLM 158
QQP P QQ Q QQQ +Q QQ AS Q T QQQ
Sbjct: 145 QQPSLFQTPGQQQASPFQTPGQQQ---SSPFQTPGQQQASPFQ------TPGQQQ----- 282
Query: 159 QQPKSMGQPLLQQQQQQLLQQQLQLQQQYQRLQQQLASSAQLQQNSSTLNQLPQLMQQQK 218
P S P QQQQQ LQ Q Q QQQ Q+LQQQ QLQQ
Sbjct: 283 PSPFSQITPFSQQQQQ--LQFQQQQQQQQQQLQQQ--QQQQLQQ---------------- 402
Query: 219 SMGQPLLQQQLQQQQLLLQQQQLVLQQQQQQQQQ 252
QQQLQ QQ QQQQL+ Q Q QQQQ
Sbjct: 403 -------QQQLQLQQ---QQQQLLQQHQSSQQQQ 474
>TC89480 similar to GP|20260684|gb|AAM13240.1 unknown protein {Arabidopsis
thaliana}, partial (43%)
Length = 1068
Score = 60.5 bits (145), Expect = 1e-09
Identities = 59/173 (34%), Positives = 79/173 (45%), Gaps = 12/173 (6%)
Frame = +1
Query: 101 PKPMGQPQLQQQLQHQQLLQQQLQLQQQYQRLQQQLASSAQLQQNSLTLNQQQLPLLMQQ 160
P P PQ+Q Q Q Q L LQQQ Q L QQ Q Q + + QQQ QQ
Sbjct: 91 PTPNSNPQIQSQSQSQSNQDPNLYLQQQQQFLHQQQQQPFQQSQTTQSQFQQQ-----QQ 255
Query: 161 PKSMGQPLLQQQQQQLL----QQQLQLQQQYQRLQQQLASS---AQLQQNSSTLNQLPQL 213
Q L QQQQQQ + QQQLQ QQ Q L Q LAS L +N + + +
Sbjct: 256 -----QQLYQQQQQQRILQQQQQQLQQPQQQQNLHQSLASHFHLLNLVENLAEVVEHGNP 420
Query: 214 MQQQKSMGQPLLQQQLQQQQLLLQ-----QQQLVLQQQQQQQQQQRTQLLQQQ 261
QQ ++ L + QQLL + + + Q+++ ++ QLL Q+
Sbjct: 421 DQQSDALITELSNHFEKCQQLLNSISASISTKAMTVEGQKKKLEESEQLLNQR 579
Score = 55.8 bits (133), Expect = 2e-08
Identities = 44/111 (39%), Positives = 52/111 (46%)
Frame = +1
Query: 157 LMQQPKSMGQPLLQQQQQQLLQQQLQLQQQYQRLQQQLASSAQLQQNSSTLNQLPQLMQQ 216
++ P S Q Q Q Q L LQQQ Q L QQ QQ+ +T +Q Q QQ
Sbjct: 85 MIPTPNSNPQIQSQSQSQSNQDPNLYLQQQQQFLHQQ--QQQPFQQSQTTQSQFQQQQQQ 258
Query: 217 QKSMGQPLLQQQLQQQQLLLQQQQLVLQQQQQQQQQQRTQLLQQQMQALRI 267
Q L QQ QQQ++L QQQQ Q QQ QQQQ Q L L +
Sbjct: 259 Q-------LYQQQQQQRILQQQQQ---QLQQPQQQQNLHQSLASHFHLLNL 381
Score = 53.9 bits (128), Expect = 9e-08
Identities = 57/190 (30%), Positives = 89/190 (46%), Gaps = 17/190 (8%)
Frame = +1
Query: 48 QNQLLKQIPAVSGSASLLPLQQPQSQQQLASSAQLQQS---SLTLNQQQLPQLMQQPKPM 104
+++ ++ + G+ +++P P S Q+ S +Q Q + +L L QQQ QQ +P
Sbjct: 37 KDRAMEHYSSPGGTWTMIPT--PNSNPQIQSQSQSQSNQDPNLYLQQQQQFLHQQQQQPF 210
Query: 105 GQPQLQQQLQHQQLLQQQLQLQQQYQRL---QQQLASSAQLQQN---SLTLNQQQLPLLM 158
Q Q Q Q QQ QQQL QQQ QR+ QQQ Q QQN SL + L L+
Sbjct: 211 QQSQTTQS-QFQQQQQQQLYQQQQQQRILQQQQQQLQQPQQQQNLHQSLASHFHLLNLVE 387
Query: 159 QQPKSMGQPLLQQQQQQLLQQQLQLQQQYQRLQQQLAS-SAQLQQNSST-------LNQL 210
+ + QQ L+ +L +++ QQ L S SA + + T L +
Sbjct: 388 NLAEVVEHGNPDQQSDALI---TELSNHFEKCQQLLNSISASISTKAMTVEGQKKKLEES 558
Query: 211 PQLMQQQKSM 220
QL+ Q++ +
Sbjct: 559 EQLLNQRRDL 588
Score = 48.5 bits (114), Expect = 4e-06
Identities = 33/74 (44%), Positives = 41/74 (54%), Gaps = 1/74 (1%)
Frame = +1
Query: 196 SSAQLQ-QNSSTLNQLPQLMQQQKSMGQPLLQQQLQQQQLLLQQQQLVLQQQQQQQQQQR 254
S+ Q+Q Q+ S NQ P L QQ+ QQQ QQ Q Q QQQQ QQQQ+
Sbjct: 103 SNPQIQSQSQSQSNQDPNLYLQQQQQFLHQQQQQPFQQSQTTQSQFQQQQQQQLYQQQQQ 282
Query: 255 TQLLQQQMQALRIP 268
++LQQQ Q L+ P
Sbjct: 283 QRILQQQQQQLQQP 324
>TC85969 similar to GP|21553908|gb|AAM62991.1 unknown {Arabidopsis thaliana},
partial (31%)
Length = 2756
Score = 59.7 bits (143), Expect = 2e-09
Identities = 51/169 (30%), Positives = 74/169 (43%)
Frame = +2
Query: 95 PQLMQQPKPMGQPQLQQQLQHQQLLQQQLQLQQQYQRLQQQLASSAQLQQNSLTLNQQQL 154
PQ++Q QPQ Q Q Q Q Q Q Q Q Q + QQ+ + +
Sbjct: 1289 PQVVQV-----QPQAQNQAQTQSQAQSQAQTQSQAPQFQQKSSGGG---------SDLPS 1426
Query: 155 PLLMQQPKSMGQPLLQQQQQQLLQQQLQLQQQYQRLQQQLASSAQLQQNSSTLNQLPQLM 214
P + S+ + +Q+ + Q+Q+Q+Q R+ + L S + Q+ Q +
Sbjct: 1427 PDSVSSDSSLSNAM-SRQKPVVYQEQIQIQPGTTRVSNPVDPKLNLSDPHSRI-QVQQHV 1600
Query: 215 QQQKSMGQPLLQQQLQQQQLLLQQQQLVLQQQQQQQQQQRTQLLQQQMQ 263
Q + Q + Q QQQQ LQQQQ Q Q QQ Q Q QQQ+Q
Sbjct: 1601 QDPGYLLQQQFELQQQQQQFELQQQQQQPQHQPQQSQPQHQPQQQQQLQ 1747
Score = 46.2 bits (108), Expect = 2e-05
Identities = 50/162 (30%), Positives = 66/162 (39%), Gaps = 2/162 (1%)
Frame = +2
Query: 21 QLQLSQQAPFNNQQLVGLPQSGQPAMIQNQLLKQIPAVSGSASLLPLQQPQSQQQLASSA 80
Q Q+ Q P Q Q+ A Q+Q + SG S LP S S+A
Sbjct: 1286 QPQVVQVQPQAQNQAQTQSQAQSQAQTQSQAPQFQQKSSGGGSDLPSPDSVSSDSSLSNA 1465
Query: 81 QLQQSSLTLNQQ-QL-PQLMQQPKPMGQPQLQQQLQHQQLLQQQLQLQQQYQRLQQQLAS 138
+Q + +Q Q+ P + P+ P+L H ++ QQ +Q LQQQ
Sbjct: 1466 MSRQKPVVYQEQIQIQPGTTRVSNPV-DPKLNLSDPHSRIQVQQ-HVQDPGYLLQQQFEL 1639
Query: 139 SAQLQQNSLTLNQQQLPLLMQQPKSMGQPLLQQQQQQLLQQQ 180
Q QQ L QQQ QQ + QP QQQ Q QQQ
Sbjct: 1640 QQQQQQFELQQQQQQPQHQPQQSQPQHQPQQQQQLQHHQQQQ 1765
Score = 43.9 bits (102), Expect = 1e-04
Identities = 55/222 (24%), Positives = 82/222 (36%), Gaps = 1/222 (0%)
Frame = +2
Query: 73 QQQLASSAQLQQSSLTLNQQQLPQLMQQPKPMGQPQLQQQLQHQQLLQQQLQLQQQYQRL 132
QQQ Q QQ L QQQ QQ +P QPQ QQQLQH Q QQQ+
Sbjct: 1622 QQQFELQQQQQQFELQQQQQQPQHQPQQSQPQHQPQQQQQLQHHQ--------QQQF--- 1768
Query: 133 QQQLASSAQLQQNSLTLNQQQLPLLMQQPKSMGQPLLQQQQQQLLQQQLQLQQQYQRLQQ 192
+ + N + QQP + +Q QQ +Q Q
Sbjct: 1769 ---IHGGHYIHHNPAIPTYYPVYPSQQQPHHQVYYVPARQPQQGYNISVQ--------QA 1915
Query: 193 QLASSAQLQQNSSTLN-QLPQLMQQQKSMGQPLLQQQLQQQQLLLQQQQLVLQQQQQQQQ 251
+ SA +S N P + QQ + P+ L + ++ + Q Q
Sbjct: 1916 NMGESATTIPSSRPQNPPNPTTLVQQNAAYNPIRSAPLPKTEMTAAAYRAATGGSPQFVQ 2095
Query: 252 QQRTQLLQQQMQALRIPGPAGHKSLSLTGSQPEATAFGMTTP 293
+Q QQ + +I P+ +S++ + P + A+ P
Sbjct: 2096 VPTSQHQQQYVTYSQIHHPS--QSMAPNSAAPASYAYEYADP 2215
>TC79570 similar to PIR|F86427|F86427 auxin response factor 6 (ARF6)
[imported] - Arabidopsis thaliana, partial (15%)
Length = 1143
Score = 57.8 bits (138), Expect = 6e-09
Identities = 51/162 (31%), Positives = 73/162 (44%), Gaps = 6/162 (3%)
Frame = +2
Query: 69 QPQSQQQLASSAQLQQSSLTLNQQQLPQLMQQPKPMGQPQLQQQLQHQQLLQQQLQLQQQ 128
QP Q +A++A +L ++Q L+Q +P L Q+LQQ Q
Sbjct: 287 QPDMYQAMAAAALQDMRTLDPSKQHPASLLQFQQPQNFSNGNAALMQNQMLQQSQPHQAF 466
Query: 129 YQRLQQQLASSAQLQQNSLTLNQQQLPLLMQQPKSMGQPL-LQQQQQQLLQQQLQLQQQY 187
+ + Q S +Q Q QQ LL Q Q +QQQQQQ QQQ Q QQQ+
Sbjct: 467 PKNQESQHPSQSQAQ------TQQFQQLLQHQHSFTNQNYHMQQQQQQQQQQQQQQQQQH 628
Query: 188 QRLQQQLASSAQL----QQNSSTLNQLPQLMQQQKSMG-QPL 224
Q+ QQQ + Q QQ S+ ++ + Q + S QP+
Sbjct: 629 QQQQQQQSQQQQQVVDHQQMSNAVSTMSQFVSAPPSQSTQPM 754
Score = 55.8 bits (133), Expect = 2e-08
Identities = 43/127 (33%), Positives = 61/127 (47%), Gaps = 2/127 (1%)
Frame = +2
Query: 132 LQQQLASSAQLQQNSLTLNQQQLPLLMQ--QPKSMGQPLLQQQQQQLLQQQLQLQQQYQR 189
+ Q +A++A +L ++Q L+Q QP++ Q Q+LQQ Q Q + +
Sbjct: 296 MYQAMAAAALQDMRTLDPSKQHPASLLQFQQPQNFSNGNAALMQNQMLQQS-QPHQAFPK 472
Query: 190 LQQQLASSAQLQQNSSTLNQLPQLMQQQKSMGQPLLQQQLQQQQLLLQQQQLVLQQQQQQ 249
Q+ S Q+ + Q QL+Q Q S Q QQQQ QQQQ Q QQQQ
Sbjct: 473 NQE----SQHPSQSQAQTQQFQQLLQHQHSFTNQNYHMQQQQQQQQQQQQQQQQQHQQQQ 640
Query: 250 QQQQRTQ 256
QQQ + Q
Sbjct: 641 QQQSQQQ 661
Score = 52.8 bits (125), Expect = 2e-07
Identities = 57/168 (33%), Positives = 73/168 (42%), Gaps = 2/168 (1%)
Frame = +2
Query: 56 PAVSGSASLLPLQQPQSQQQLASSAQLQQSSLTLNQ--QQLPQLMQQPKPMGQPQLQQQL 113
P+ ASLL QQPQ+ ++A +Q L +Q Q P+ + P Q Q Q Q
Sbjct: 347 PSKQHPASLLQFQQPQNFSN-GNAALMQNQMLQQSQPHQAFPKNQESQHP-SQSQAQTQ- 517
Query: 114 QHQQLLQQQLQLQQQYQRLQQQLASSAQLQQNSLTLNQQQLPLLMQQPKSMGQPLLQQQQ 173
Q QQLLQ Q Q +QQQ Q QQ +QQQ QQ +S QQQ
Sbjct: 518 QFQQLLQHQHSFTNQNYHMQQQQQQQQQQQQQQQQQHQQQ-----QQQQS-------QQQ 661
Query: 174 QQLLQQQLQLQQQYQRLQQQLASSAQLQQNSSTLNQLPQLMQQQKSMG 221
QQ++ Q Q A S Q S+ +Q Q MQ S+G
Sbjct: 662 QQVVDHQ----------QMSNAVSTMSQFVSAPPSQSTQPMQAISSIG 775
Score = 32.3 bits (72), Expect = 0.29
Identities = 54/216 (25%), Positives = 73/216 (33%), Gaps = 13/216 (6%)
Frame = +2
Query: 20 QQLQLSQQAPFNNQQLVGLPQSGQPAMIQNQLLKQIPAVSGSASLLPLQQPQSQQQLASS 79
QQ Q Q P N + Q Q L Q + + + QQ Q QQQ
Sbjct: 440 QQSQPHQAFPKNQESQHPSQSQAQTQQFQQLLQHQHSFTNQNYHMQQQQQQQQQQQ---- 607
Query: 80 AQLQQSSLTLNQQQLPQLMQQPKPMGQPQLQQQLQHQQL------LQQQLQLQQQYQRLQ 133
QQQ Q QQ + Q Q QQ + HQQ+ + Q +
Sbjct: 608 -----------QQQQQQHQQQQQQQSQQQ-QQVVDHQQMSNAVSTMSQFVSAPPSQSTQP 751
Query: 134 QQLASSAQLQQNSLTLNQQQLPLLMQQPKSMGQPLLQQQQQQLL-----QQQLQLQQQYQ 188
Q SS QQN N + L S + LL + +Q
Sbjct: 752 MQAISSIGHQQNFSDSNGNPVSPLHNMLGSFS----NDETSHLLNFPRPNSWVPVQSSTA 919
Query: 189 RLQQQLASSAQLQQNSS--TLNQLPQLMQQQKSMGQ 222
R +++A L +S L Q+ QL Q Q +M Q
Sbjct: 920 RPSKRVAVDPLLSSGASHYVLPQVEQLGQSQTTMSQ 1027
>TC91076 similar to PIR|H96747|H96747 unknown protein T10D10.14 [imported] -
Arabidopsis thaliana, partial (9%)
Length = 834
Score = 56.6 bits (135), Expect = 1e-08
Identities = 70/239 (29%), Positives = 96/239 (39%), Gaps = 4/239 (1%)
Frame = +3
Query: 35 LVGLPQSGQPAMIQNQLLKQIPAVSGSASLLPLQQPQSQQQLASSAQLQQSSLTLNQQQL 94
LV LP Q ++ N++ + + G QS + A A +SL L Q +
Sbjct: 228 LVNLPLGSQSSLPPNEMQQYGEPIPG----------QSNNEAAKLAGGSNASLNLPQSLV 377
Query: 95 PQLMQQPKPMGQPQLQQQLQHQQLLQQQLQLQQQYQRLQQQLASSAQLQQNSLTLNQQQL 154
P G PQ LQ Q L + + Q+ Q+L Q
Sbjct: 378 ANARMLPP--GNPQ---GLQMSQALLSGVSMAQRPQQLDSQ------------------- 485
Query: 155 PLLMQQPKSMGQPLLQQQQQQLLQQQLQLQQQYQRLQQQLASSAQLQQNSSTLNQLPQL- 213
Q +LQQQQQQ QQLQ Q Q+ LQQQ + Q Q++ + NQL L
Sbjct: 486 -----------QAVLQQQQQQ---QQLQ-QNQHSLLQQQ---NPQFQRSLLSANQLSHLN 611
Query: 214 ---MQQQKSMGQPLLQQQLQQQQLLLQQQQLVLQQQQQQQQQQRTQLLQQQMQALRIPG 269
+G LL + Q +LQQQ QQQQ QQ Q+ Q+ ++ M L G
Sbjct: 612 GVGQNSNMPLGNHLLNKASPLQIQMLQQQH----QQQQLQQNQQPQMQRKMMMGLGAMG 776
Score = 39.7 bits (91), Expect = 0.002
Identities = 46/176 (26%), Positives = 67/176 (37%), Gaps = 7/176 (3%)
Frame = +3
Query: 108 QLQQQLQHQQLLQQQLQLQQQYQRLQQQLASSAQLQQNSLTLNQQQLPLLMQQPKSMGQP 167
Q Q++H +++ ++Q + + L S + L N MQQ G+P
Sbjct: 159 QFSSQIEHDGYVKEDDRIQVRPNLVNLPLGSQSSLPPNE-----------MQQ---YGEP 296
Query: 168 LLQQQQQQLLQQQLQLQQQYQRLQQQLASSAQLQQNSSTLNQLPQLMQQQKSMGQPLLQQ 227
+ Q + +A+L S+ LPQ + M P Q
Sbjct: 297 IPGQSNNE---------------------AAKLAGGSNASLNLPQSLVANARMLPPGNPQ 413
Query: 228 QLQQQQLLL-------QQQQLVLQQQQQQQQQQRTQLLQQQMQALRIPGPAGHKSL 276
LQ Q LL + QQL QQ QQQQQ+ QL Q Q L+ P +SL
Sbjct: 414 GLQMSQALLSGVSMAQRPQQLDSQQAVLQQQQQQQQLQQNQHSLLQQQNPQFQRSL 581
>TC92665 homologue to GP|15291583|gb|AAK93060.1 GH28569p {Drosophila
melanogaster}, partial (4%)
Length = 668
Score = 39.7 bits (91), Expect = 0.002
Identities = 28/82 (34%), Positives = 40/82 (48%)
Frame = +3
Query: 41 SGQPAMIQNQLLKQIPAVSGSASLLPLQQPQSQQQLASSAQLQQSSLTLNQQQLPQLMQQ 100
S Q ++I Q V S S LP Q + Q ++ ++LQQ S + QQQ ++ Q
Sbjct: 18 SSQQSVIPTSSAMQPSMVQSSLSSLPQNQQSNNVQQSTQSRLQQHSQIIRQQQQNSIVNQ 197
Query: 101 PKPMGQPQLQQQLQHQQLLQQQ 122
+ P +QQ QHQQL Q
Sbjct: 198 QQ---TPMIQQ--QHQQLAGPQ 248
Score = 38.9 bits (89), Expect(2) = 3e-07
Identities = 33/93 (35%), Positives = 44/93 (46%)
Frame = +3
Query: 75 QLASSAQLQQSSLTLNQQQLPQLMQQPKPMGQPQLQQQLQHQQLLQQQLQLQQQYQRLQQ 134
QL SS QQS + + P ++Q PQ QQ QQ Q +LQ Q R
Sbjct: 9 QLLSS---QQSVIPTSSAMQPSMVQSSLS-SLPQNQQSNNVQQSTQSRLQQHSQIIR--- 167
Query: 135 QLASSAQLQQNSLTLNQQQLPLLMQQPKSMGQP 167
Q QQNS+ +NQQQ P++ QQ + + P
Sbjct: 168 ------QQQQNSI-VNQQQTPMIQQQHQQLAGP 245
Score = 38.5 bits (88), Expect = 0.004
Identities = 28/80 (35%), Positives = 41/80 (51%), Gaps = 2/80 (2%)
Frame = +3
Query: 191 QQQLASSAQLQQNSSTLNQLPQLMQQQKSMGQPLLQQQLQQQQLLLQQQQLVLQQQQQQQ 250
QQ + ++ Q S + L L Q Q+S +QQ Q + LQQ +++QQQQ
Sbjct: 24 QQSVIPTSSAMQPSMVQSSLSSLPQNQQSNN---VQQSTQSR---LQQHSQIIRQQQQNS 185
Query: 251 --QQQRTQLLQQQMQALRIP 268
QQ+T ++QQQ Q L P
Sbjct: 186 IVNQQQTPMIQQQHQQLAGP 245
Score = 37.4 bits (85), Expect = 0.009
Identities = 28/75 (37%), Positives = 37/75 (49%), Gaps = 6/75 (8%)
Frame = +1
Query: 105 GQPQLQQQLQHQQLLQQQ-----LQLQQQYQRLQQQLASSAQLQQNSLTLNQQQ-LPLLM 158
G Q+Q+ QH Q+L QQ +Q Q+ Q L + Q QQ +N Q L +
Sbjct: 241 GLSQMQRNGQHAQMLGQQNNVGDVQKSQRLHPQQNNLMNLQQRQQQQQLMNHQNNLTNIH 420
Query: 159 QQPKSMGQPLLQQQQ 173
QQP + Q L QQQQ
Sbjct: 421 QQPGNNVQGLQQQQQ 465
Score = 35.0 bits (79), Expect = 0.045
Identities = 27/73 (36%), Positives = 38/73 (51%), Gaps = 3/73 (4%)
Frame = +3
Query: 195 ASSAQLQQNSSTLNQLPQLMQQQKSMGQPLLQQQLQQQQLLLQQQQL--VLQQQQQQQ-Q 251
+S+ Q S+L+ LPQ QQ + Q Q +LQQ +++QQQ ++ QQQ Q
Sbjct: 45 SSAMQPSMVQSSLSSLPQ--NQQSNNVQQSTQSRLQQHSQIIRQQQQNSIVNQQQTPMIQ 218
Query: 252 QQRTQLLQQQMQA 264
QQ QL Q A
Sbjct: 219 QQHQQLAGPQSNA 257
Score = 32.7 bits (73), Expect = 0.22
Identities = 24/77 (31%), Positives = 38/77 (49%), Gaps = 2/77 (2%)
Frame = +3
Query: 175 QLLQQQLQLQQQYQRLQQQLASSA--QLQQNSSTLNQLPQLMQQQKSMGQPLLQQQLQQQ 232
QLL Q + +Q + S+ L QN + N + Q Q + +++QQ QQ
Sbjct: 9 QLLSSQQSVIPTSSAMQPSMVQSSLSSLPQNQQS-NNVQQSTQSRLQQHSQIIRQQ-QQN 182
Query: 233 QLLLQQQQLVLQQQQQQ 249
++ QQQ ++QQQ QQ
Sbjct: 183 SIVNQQQTPMIQQQHQQ 233
Score = 32.7 bits (73), Expect(2) = 3e-07
Identities = 29/96 (30%), Positives = 43/96 (44%)
Frame = +1
Query: 194 LASSAQLQQNSSTLNQLPQLMQQQKSMGQPLLQQQLQQQQLLLQQQQLVLQQQQQQQQQQ 253
L +Q+Q+N Q Q++ QQ ++G +Q+ Q L QQ ++ QQ+QQQQ
Sbjct: 235 LRGLSQMQRNG----QHAQMLGQQNNVGD------VQKSQRLHPQQNNLMNLQQRQQQQ- 381
Query: 254 RTQLLQQQMQALRIPGPAGHKSLSLTGSQPEATAFG 289
QL+ Q I G+ L Q T G
Sbjct: 382 --QLMNHQNNLTNIHQQPGNNVQGLQQQQQFGTESG 483
Score = 28.1 bits (61), Expect = 5.5
Identities = 30/120 (25%), Positives = 44/120 (36%), Gaps = 6/120 (5%)
Frame = +1
Query: 59 SGSASLLPLQQPQSQQQLASSAQLQQSSLTLNQQQLPQLMQQPKPMGQPQLQQQLQHQQL 118
S +SL L Q Q Q A Q + + + Q P+ LQQ+ Q QQL
Sbjct: 220 SNISSLRGLSQMQRNGQHAQMLGQQNNVGDVQKSQ----RLHPQQNNLMNLQQRQQQQQL 387
Query: 119 LQQQLQLQQQYQRLQQQLASSAQLQQNSLTLNQQQLPL------LMQQPKSMGQPLLQQQ 172
+ Q L +Q+ + Q QQ Q + ++QQPK + Q
Sbjct: 388 MNHQNNLTNIHQQPGNNVQGLQQQQQFGTESGNQGIQTSHHSAQMLQQPKGFNAATIATQ 567
Score = 27.7 bits (60), Expect = 7.1
Identities = 28/91 (30%), Positives = 39/91 (42%), Gaps = 2/91 (2%)
Frame = +3
Query: 117 QLLQQQLQLQQQYQRLQQQLASSA--QLQQNSLTLNQQQLPLLMQQPKSMGQPLLQQQQQ 174
QLL Q + +Q + S+ L QN + N QQ Q S Q + QQQQ
Sbjct: 9 QLLSSQQSVIPTSSAMQPSMVQSSLSSLPQNQQSNNVQQSTQSRLQQHS--QIIRQQQQN 182
Query: 175 QLLQQQLQLQQQYQRLQQQLASSAQLQQNSS 205
++ QQ Q +QQQ A Q N++
Sbjct: 183 SIVNQQ-----QTPMIQQQHQQLAGPQSNAT 260
>TC88336 similar to PIR|T51477|T51477 glutamine-rich protein - Arabidopsis
thaliana, partial (67%)
Length = 1297
Score = 50.8 bits (120), Expect = 8e-07
Identities = 43/122 (35%), Positives = 62/122 (50%), Gaps = 17/122 (13%)
Frame = +2
Query: 84 QSSLTLNQQQLPQLMQQPKPMGQPQLQQQLQHQQLLQQQLQLQQQYQRLQQQLASSAQLQ 143
+ + L+QQQ Q QQ + + Q Q +QQ Q +L QQLQ QQQ QQQ A+ ++
Sbjct: 176 EEAKALHQQQQQQQQQQQQLLIQEQQRQQQQQHFMLLQQLQKQQQQ---QQQAAAISRFP 346
Query: 144 QN-SLTLNQQQLPLLMQQ---PKSMGQPLL-----------QQQQQQLLQ--QQLQLQQQ 186
N L + PL +QQ P P+L QQQQQ++++ Q++LQ
Sbjct: 347 SNIDAHLRPIRPPLNLQQNPNPNPNSNPILNLHQNPNSNHQQQQQQKMIRPGNQMELQMA 526
Query: 187 YQ 188
YQ
Sbjct: 527 YQ 532
Score = 45.8 bits (107), Expect = 3e-05
Identities = 36/108 (33%), Positives = 51/108 (46%), Gaps = 11/108 (10%)
Frame = +2
Query: 158 MQQPKSMGQPLLQQQQQQ----LLQQQLQLQQQYQRLQQQLASSAQLQQNSSTLNQLPQL 213
M++ K++ Q QQQQQQ + +QQ Q QQQ+ L QQL Q QQ ++ +++ P
Sbjct: 173 MEEAKALHQQQQQQQQQQQQLLIQEQQRQQQQQHFMLLQQLQKQQQQQQQAAAISRFPSN 352
Query: 214 MQQQKSMGQPLLQQQLQQQQ-------LLLQQQQLVLQQQQQQQQQQR 254
+ +P L Q L L Q QQQQQQ+ R
Sbjct: 353 IDAHLRPIRPPLNLQQNPNPNPNSNPILNLHQNPNSNHQQQQQQKMIR 496
Score = 45.4 bits (106), Expect = 3e-05
Identities = 26/54 (48%), Positives = 33/54 (60%)
Frame = +2
Query: 214 MQQQKSMGQPLLQQQLQQQQLLLQQQQLVLQQQQQQQQQQRTQLLQQQMQALRI 267
M++ K++ Q QQQ QQQQLL+Q+QQ QQQ QQ + QQQ QA I
Sbjct: 173 MEEAKALHQQQQQQQQQQQQLLIQEQQRQQQQQHFMLLQQLQKQQQQQQQAAAI 334
Score = 40.4 bits (93), Expect = 0.001
Identities = 42/134 (31%), Positives = 59/134 (43%), Gaps = 14/134 (10%)
Frame = +2
Query: 129 YQRLQQQLASSAQLQQNSLTLNQQQLPLLMQQPKSMGQPLLQQQQQQLLQQQLQLQQQYQ 188
+ R ++++ + L Q QQQ LL+Q+ Q QQQQ +L QQLQ QQ Q
Sbjct: 152 FSRFRKKMEEAKALHQQQQQQQQQQQQLLIQE-----QQRQQQQQHFMLLQQLQKQQ--Q 310
Query: 189 RLQQQLASSAQLQQNSSTLNQL-PQLMQQQ----KSMGQPLL---------QQQLQQQQL 234
+ QQ A S + L + P L QQ P+L QQ QQQ++
Sbjct: 311 QQQQAAAISRFPSNIDAHLRPIRPPLNLQQNPNPNPNSNPILNLHQNPNSNHQQQQQQKM 490
Query: 235 LLQQQQLVLQQQQQ 248
+ Q+ LQ Q
Sbjct: 491 IRPGNQMELQMAYQ 532
Score = 31.6 bits (70), Expect = 0.49
Identities = 19/43 (44%), Positives = 26/43 (60%), Gaps = 8/43 (18%)
Frame = +2
Query: 226 QQQLQQQQLLLQQQQLVLQQQQQ--------QQQQQRTQLLQQ 260
++++++ + L QQQQ QQQQQ QQQQQ LLQQ
Sbjct: 164 RKKMEEAKALHQQQQQQQQQQQQLLIQEQQRQQQQQHFMLLQQ 292
Score = 30.4 bits (67), Expect = 1.1
Identities = 24/83 (28%), Positives = 40/83 (47%), Gaps = 1/83 (1%)
Frame = +2
Query: 187 YQRLQQQLASSAQLQQNSSTLNQLPQLMQQQKSMGQPLLQQQLQQQQLLLQQ-QQLVLQQ 245
+ R ++++ + L Q Q Q QQQ+ + Q +QQ QQ +LLQQ Q+ QQ
Sbjct: 152 FSRFRKKMEEAKALHQQ-----QQQQQQQQQQLLIQEQQRQQQQQHFMLLQQLQKQQQQQ 316
Query: 246 QQQQQQQQRTQLLQQQMQALRIP 268
QQ + + ++ +R P
Sbjct: 317 QQAAAISRFPSNIDAHLRPIRPP 385
>BG586394 similar to GP|7295463|gb| CG4835 gene product {Drosophila
melanogaster}, partial (2%)
Length = 734
Score = 50.8 bits (120), Expect = 8e-07
Identities = 61/203 (30%), Positives = 96/203 (47%), Gaps = 24/203 (11%)
Frame = +2
Query: 87 LTLNQQQLPQLMQQPK--PMGQPQLQQQLQHQ--------QLLQQQLQL----QQQYQRL 132
L NQ++ L Q + P + Q+ Q+L Q LL+ Q Q Q+Q R
Sbjct: 14 LLRNQKKRLLLRSQKQILPRNRKQMFQRLPRQ*KKVKMILGLLRNQKQRHPRNQKQMFRK 193
Query: 133 QQQLASSAQLQQNSLTLNQQQLPLLMQQPKSMGQPLLQQQQQQLLQQQLQLQQQYQRLQQ 192
QQQL +L Q L Q+ LL Q+ ++ Q LL+ Q+Q L + Q Q+ Q+ R +
Sbjct: 194 QQQLNLLFKLIQKKHQLKNQKKRLL-QKLQNQKQRLLRNQKQILPRNQKQMFQRLLRH*K 370
Query: 193 QLASSAQLQQNSSTLNQLPQLMQQQKSMGQPLLQQQLQQQQLLLQQQQLVLQQQ------ 246
++ L +N + Q +QQ+ + + L+ Q Q Q LLQ Q+ + QQQ
Sbjct: 371 KIKMFPDLLRNQKQRHPRNQKRKQQQ-LKRHQLKNQKQIFQRLLQHQKKIKQQQLNLLFQ 547
Query: 247 ----QQQQQQQRTQLLQQQMQAL 265
+ Q + Q+ +LL+ Q Q L
Sbjct: 548 LFQKRHQPKNQKKRLLRHQKQRL 616
Score = 30.8 bits (68), Expect = 0.84
Identities = 45/158 (28%), Positives = 65/158 (40%), Gaps = 17/158 (10%)
Frame = +2
Query: 20 QQLQLSQQAPFNNQQLVGLPQSGQPAMIQNQL-----LKQIP-AVSGSASLLPLQQPQSQ 73
Q+LQ +Q NQ+ + LP++ Q M Q L +K P + P Q + Q
Sbjct: 269 QKLQNQKQRLLRNQKQI-LPRN-QKQMFQRLLRH*KKIKMFPDLLRNQKQRHPRNQKRKQ 442
Query: 74 QQLASSA---------QLQQSSLTLNQQQLPQLMQQPKPMGQPQLQQQLQHQQLLQQQLQ 124
QQL +L Q + QQQL L Q Q +HQ Q++
Sbjct: 443 QQLKRHQLKNQKQIFQRLLQHQKKIKQQQLNLLFQ----------LFQKRHQPKNQKKRL 592
Query: 125 LQQQYQRL--QQQLASSAQLQQNSLTLNQQQLPLLMQQ 160
L+ Q QRL Q+L ++Q+ Q+L LL Q
Sbjct: 593 LRHQKQRLPHNQKLIIPPRIQKKIRLPRHQKLRLLRHQ 706
>NP212732 NP212732|AF106929.1|AAD39890.1 putative cell wall protein
Length = 576
Score = 50.1 bits (118), Expect = 1e-06
Identities = 41/147 (27%), Positives = 66/147 (44%), Gaps = 2/147 (1%)
Frame = +2
Query: 108 QLQQQLQHQQLLQQQLQLQQQYQRLQQQLASSAQLQQNSLTLNQQQLPLLMQQP--KSMG 165
Q Q +H Q++ L+++ +++ QL Q Q Q Q+ L + P K +
Sbjct: 113 QQNQHQEHHHHHQKERMLKERIHQMEMQLKERMQHQMELKEKMQHQMELKERMPLKKELR 292
Query: 166 QPLLQQQQQQLLQQQLQLQQQYQRLQQQLASSAQLQQNSSTLNQLPQLMQQQKSMGQPLL 225
+ L Q QQ Q Q Q Q+Q+Q + +Q Q Q+K LL
Sbjct: 293 EL*LHQHQQHQEQHQEQHQEQHQHQEDH--------------HQKEQHHHQRKEEQPHLL 430
Query: 226 QQQLQQQQLLLQQQQLVLQQQQQQQQQ 252
Q Q Q++QLL Q++Q +QQQ+ +
Sbjct: 431 QVQEQERQLLQQERQEAQLPKQQQEPE 511
>TC92466 similar to GP|11141605|gb|AAG32022.1 LEUNIG {Arabidopsis thaliana},
partial (14%)
Length = 632
Score = 49.3 bits (116), Expect = 2e-06
Identities = 40/96 (41%), Positives = 44/96 (45%)
Frame = +1
Query: 170 QQQQQQLLQQQLQLQQQYQRLQQQLASSAQLQQNSSTLNQLPQLMQQQKSMGQPLLQQQL 229
+QQQQQ QQQ QQ Q QQQ Q+QQ LMQ+ Q QQQ
Sbjct: 370 EQQQQQQQQQQQPQPQQSQHAQQQQQQHMQMQQ---------LLMQRHAQQQQ---QQQQ 513
Query: 230 QQQQLLLQQQQLVLQQQQQQQQQQRTQLLQQQMQAL 265
QQQ Q QQ Q QQQQ + RT LL L
Sbjct: 514 HQQQPQSQPQQ---PQPQQQQNRDRTHLLNGSANGL 612
Score = 45.1 bits (105), Expect = 4e-05
Identities = 36/94 (38%), Positives = 45/94 (47%)
Frame = +1
Query: 178 QQQLQLQQQYQRLQQQLASSAQLQQNSSTLNQLPQLMQQQKSMGQPLLQQQLQQQQLLLQ 237
+QQ Q QQQ Q+ Q Q + AQ QQ QQ M Q L+Q+ QQQQ
Sbjct: 370 EQQQQQQQQQQQPQPQQSQHAQQQQ------------QQHMQMQQLLMQRHAQQQQ---- 501
Query: 238 QQQLVLQQQQQQQQQQRTQLLQQQMQALRIPGPA 271
QQQ QQ Q Q QQ + Q Q + + + G A
Sbjct: 502 QQQQHQQQPQSQPQQPQPQQQQNRDRTHLLNGSA 603
Score = 45.1 bits (105), Expect = 4e-05
Identities = 32/72 (44%), Positives = 38/72 (52%), Gaps = 5/72 (6%)
Frame = +1
Query: 91 QQQLPQLMQQPKPM----GQPQLQQQLQHQQLL-QQQLQLQQQYQRLQQQLASSAQLQQN 145
QQQ Q QQP+P Q Q QQ +Q QQLL Q+ Q QQQ Q+ QQQ S Q Q
Sbjct: 376 QQQQQQQQQQPQPQQSQHAQQQQQQHMQMQQLLMQRHAQQQQQQQQHQQQPQSQPQQPQP 555
Query: 146 SLTLNQQQLPLL 157
N+ + LL
Sbjct: 556 QQQQNRDRTHLL 591
Score = 43.9 bits (102), Expect = 1e-04
Identities = 33/74 (44%), Positives = 39/74 (52%)
Frame = +1
Query: 120 QQQLQLQQQYQRLQQQLASSAQLQQNSLTLNQQQLPLLMQQPKSMGQPLLQQQQQQLLQQ 179
+QQ Q QQQ Q+ Q Q + AQ QQ + Q LLMQ+ QQQQQQ QQ
Sbjct: 370 EQQQQQQQQQQQPQPQQSQHAQQQQQQ---HMQMQQLLMQRHAQ------QQQQQQQHQQ 522
Query: 180 QLQLQQQYQRLQQQ 193
Q Q Q Q + QQQ
Sbjct: 523 QPQSQPQQPQPQQQ 564
Score = 42.4 bits (98), Expect = 3e-04
Identities = 28/67 (41%), Positives = 34/67 (49%)
Frame = +1
Query: 151 QQQLPLLMQQPKSMGQPLLQQQQQQLLQQQLQLQQQYQRLQQQLASSAQLQQNSSTLNQL 210
QQQ QQP+ QQQQQQ +Q Q L Q++ + QQQ Q QQ + Q
Sbjct: 376 QQQQQQQQQQPQPQQSQHAQQQQQQHMQMQQLLMQRHAQQQQQ---QQQHQQQPQSQPQQ 546
Query: 211 PQLMQQQ 217
PQ QQQ
Sbjct: 547 PQPQQQQ 567
Score = 41.6 bits (96), Expect = 5e-04
Identities = 32/79 (40%), Positives = 35/79 (43%)
Frame = +1
Query: 68 QQPQSQQQLASSAQLQQSSLTLNQQQLPQLMQQPKPMGQPQLQQQLQHQQLLQQQLQLQQ 127
+Q Q QQQ Q QQS QQQ Q MQ + + Q QQQ Q QQ QQ Q
Sbjct: 370 EQQQQQQQQQQQPQPQQSQHAQQQQQ--QHMQMQQLLMQRHAQQQQQQQQHQQQPQSQPQ 543
Query: 128 QYQRLQQQLASSAQLQQNS 146
Q Q QQQ L S
Sbjct: 544 QPQPQQQQNRDRTHLLNGS 600
Score = 41.2 bits (95), Expect = 6e-04
Identities = 28/65 (43%), Positives = 30/65 (46%)
Frame = +1
Query: 110 QQQLQHQQLLQQQLQLQQQYQRLQQQLASSAQLQQNSLTLNQQQLPLLMQQPKSMGQPLL 169
QQQ Q QQ Q Q Q Q Q+ QQQ QL QQQ QQP+S Q
Sbjct: 373 QQQQQQQQQQQPQPQQSQHAQQQQQQHMQMQQLLMQRHAQQQQQQQQHQQQPQSQPQQPQ 552
Query: 170 QQQQQ 174
QQQQ
Sbjct: 553 PQQQQ 567
Score = 35.0 bits (79), Expect = 0.045
Identities = 28/65 (43%), Positives = 32/65 (49%), Gaps = 8/65 (12%)
Frame = +1
Query: 207 LNQLPQLMQQQKSMGQPLLQQQLQQQQLLLQQ------QQLVLQQ--QQQQQQQQRTQLL 258
LN L QQQ+ Q Q Q Q QQ QQL++Q+ QQQQQQQQ Q
Sbjct: 349 LN*LKAREQQQQQQQQQQQPQPQQSQHAQQQQQQHMQMQQLLMQRHAQQQQQQQQHQQQP 528
Query: 259 QQQMQ 263
Q Q Q
Sbjct: 529 QSQPQ 543
Score = 33.9 bits (76), Expect = 0.099
Identities = 29/73 (39%), Positives = 33/73 (44%)
Frame = +1
Query: 68 QQPQSQQQLASSAQLQQSSLTLNQQQLPQLMQQPKPMGQPQLQQQLQHQQLLQQQLQLQQ 127
QQPQ QQ + Q QQ + QQL LMQ+ Q Q Q Q Q Q QQ QQ
Sbjct: 400 QQPQPQQSQHAQQQQQQH---MQMQQL--LMQRHAQQQQQQQQHQQQPQSQPQQPQPQQQ 564
Query: 128 QYQRLQQQLASSA 140
Q + L SA
Sbjct: 565 QNRDRTHLLNGSA 603
Score = 29.6 bits (65), Expect = 1.9
Identities = 26/69 (37%), Positives = 26/69 (37%)
Frame = +1
Query: 20 QQLQLSQQAPFNNQQLVGLPQSGQPAMIQNQLLKQIPAVSGSASLLPLQQPQSQQQLASS 79
QQ Q QQ P QQ Q Q M QLL Q A QQPQSQ Q
Sbjct: 379 QQQQQQQQQP-QPQQSQHAQQQQQQHMQMQQLLMQRHAQQQQQQQQHQQQPQSQPQQPQP 555
Query: 80 AQLQQSSLT 88
Q Q T
Sbjct: 556 QQQQNRDRT 582
Score = 28.5 bits (62), Expect = 4.2
Identities = 25/72 (34%), Positives = 27/72 (36%)
Frame = +1
Query: 48 QNQLLKQIPAVSGSASLLPLQQPQSQQQLASSAQLQQSSLTLNQQQLPQLMQQPKPMGQP 107
Q Q + P S A Q Q QQ L QQ +QQQ QQP QP
Sbjct: 388 QQQQQQPQPQQSQHAQQQQQQHMQMQQLLMQRHAQQQQQQQQHQQQPQSQPQQP----QP 555
Query: 108 QLQQQLQHQQLL 119
Q QQ LL
Sbjct: 556 QQQQNRDRTHLL 591
>TC90558 similar to GP|9758584|dbj|BAB09197.1
gb|AAC80623.1~gene_id:MBD2.15~similar to unknown protein
{Arabidopsis thaliana}, partial (5%)
Length = 1047
Score = 49.3 bits (116), Expect = 2e-06
Identities = 53/157 (33%), Positives = 68/157 (42%)
Frame = +1
Query: 47 IQNQLLKQIPAVSGSASLLPLQQPQSQQQLASSAQLQQSSLTLNQQQLPQLMQQPKPMGQ 106
+ +Q Q+P L P Q LA +A SSLT + L Q P+ M
Sbjct: 604 LHDQNFPQMPFGIQQQRLTPQNQLSLSNLLAPAADNPSSSLTAEKLLSSGLSQDPQIMN- 780
Query: 107 PQLQQQLQHQQLLQQQLQLQQQYQRLQQQLASSAQLQQNSLTLNQQQLPLLMQQPKSMGQ 166
LLQQQ LQ L Q A+SA L L + L + QQ + Q
Sbjct: 781 -----------LLQQQYLLQ-----LHSQAAASAP----QLPLLDKLLLMKQQQKQEEQQ 900
Query: 167 PLLQQQQQQLLQQQLQLQQQYQRLQQQLASSAQLQQN 203
LL+QQQQQLL + LQ QQ Q +S QLQ++
Sbjct: 901 LLLRQQQQQLLSKMLQDQQSNQLFGN--SSYGQLQKS 1005
Score = 44.3 bits (103), Expect = 7e-05
Identities = 55/171 (32%), Positives = 71/171 (41%)
Frame = +1
Query: 84 QSSLTLNQQQLPQLMQQPKPMGQPQLQQQLQHQQLLQQQLQLQQQYQRLQQQLASSAQLQ 143
Q L L Q ++ L Q PQ+ +Q Q+L Q QL L LA +A
Sbjct: 565 QGGLDLLQNKIDPLHDQ----NFPQMPFGIQQQRLTPQN-QLS-----LSNLLAPAADNP 714
Query: 144 QNSLTLNQQQLPLLMQQPKSMGQPLLQQQQQQLLQQQLQLQQQYQRLQQQLASSAQLQQN 203
+SLT + L Q P+ M LLQQQ LQ L Q A+SA
Sbjct: 715 SSSLTAEKLLSSGLSQDPQIMN----------LLQQQYLLQ-----LHSQAAASA----- 834
Query: 204 SSTLNQLPQLMQQQKSMGQPLLQQQLQQQQLLLQQQQLVLQQQQQQQQQQR 254
PQL K + Q+Q +QQ LL QQQQ +L + Q QQ +
Sbjct: 835 -------PQLPLLDKLLLMKQQQKQEEQQLLLRQQQQQLLSKMLQDQQSNQ 966
Score = 41.2 bits (95), Expect = 6e-04
Identities = 43/125 (34%), Positives = 60/125 (47%), Gaps = 3/125 (2%)
Frame = +1
Query: 144 QNSLTLNQQQLPLLMQQPKSMGQPLLQQQQQQLLQQQLQLQQQYQRLQQQLASSAQLQQN 203
Q L L Q ++ L Q P QQQ+ Q QL L +SS ++
Sbjct: 565 QGGLDLLQNKIDPLHDQNFPQ-MPFGIQQQRLTPQNQLSLSNLLAPAADNPSSSLTAEKL 741
Query: 204 -SSTLNQLPQLMQ--QQKSMGQPLLQQQLQQQQLLLQQQQLVLQQQQQQQQQQRTQLLQQ 260
SS L+Q PQ+M QQ+ + Q Q QL L + L+L +QQQ+Q++Q+ L QQ
Sbjct: 742 LSSGLSQDPQIMNLLQQQYLLQLHSQAAASAPQLPLLDK-LLLMKQQQKQEEQQLLLRQQ 918
Query: 261 QMQAL 265
Q Q L
Sbjct: 919 QQQLL 933
>TC85294 similar to GP|2961298|emb|CAA12357.1 hypothetical protein {Cicer
arietinum}, partial (61%)
Length = 1353
Score = 49.3 bits (116), Expect = 2e-06
Identities = 44/153 (28%), Positives = 77/153 (49%), Gaps = 9/153 (5%)
Frame = +3
Query: 122 QLQLQQQYQRLQQQLASSAQLQQNSLTL-NQQQLPLLMQQPKSMGQPLLQQQQQQLLQ-- 178
Q Q +Q++R +Q Q++ L N ++L L Q+ + QP QQ+QQ+ LQ
Sbjct: 300 QQQCPKQHRR*SRQRKQFQNKQRSKFLLQNNRKLKFLPQKYQQKKQPNSQQKQQKKLQLP 479
Query: 179 --QQLQLQQQYQRLQQQLASSAQLQQNSSTLNQLPQ----LMQQQKSMGQPLLQQQLQQQ 232
++QL+ + ++ Q+ + NQL + L Q +S+ QPL Q QQ
Sbjct: 480 *KHKIQLKLRPRKW*QKKPRKRIQKHLKKQRNQLKK*RKRLQQ*WRSL*QPLKMSQKHQQ 659
Query: 233 QLLLQQQQLVLQQQQQQQQQQRTQLLQQQMQAL 265
Q LL+++ L Q + QQQ++ QL +++ +
Sbjct: 660 Q*LLRKRMLNQLSQWKLQQQKKFQLRNLKLKTM 758
>TC78816 similar to GP|14190377|gb|AAK55669.1 AT4g00850/A_TM018A10_22
{Arabidopsis thaliana}, partial (43%)
Length = 1150
Score = 48.9 bits (115), Expect = 3e-06
Identities = 50/184 (27%), Positives = 75/184 (40%), Gaps = 19/184 (10%)
Frame = +1
Query: 117 QLLQQQLQLQQQYQRLQQ--QLASSAQLQQNSLTLNQQQLPLLMQQPKSMGQPLLQQQQQ 174
QL Q +++ + +++QQ Q+ + +Q+ + + K + +L Q
Sbjct: 154 QLNQNPVKINNKEEKMQQTPQMIPMMPSFPQQTNITTEQIQKYLDENKKLILAILDNQNL 333
Query: 175 QLLQQQLQLQQQYQRLQQQLASSAQLQQNSSTLNQLPQL-----MQQQKSMGQPLLQQQL 229
L + Q Q Q Q+ LA+ A Q + L PQ+ MQQ M P
Sbjct: 334 GKLAECAQYQAQLQKNLMYLAAIADAQPQTPALP--PQMAPHPAMQQGFYMQHPQAAAMA 507
Query: 230 QQQQLLLQQQQLVL----QQQQQQQQQQRTQLLQQQMQALRIPGPAG--------HKSLS 277
QQQ + Q+ + Q Q QQ QQQ+ QL QQ MQ P G H +
Sbjct: 508 QQQGMFPQKMPMQFGNPHQMQDQQHQQQQQQLHQQAMQGQMGLRPGGINNGMHPMHNEAA 687
Query: 278 LTGS 281
L GS
Sbjct: 688 LGGS 699
Score = 37.0 bits (84), Expect = 0.012
Identities = 37/116 (31%), Positives = 46/116 (38%), Gaps = 7/116 (6%)
Frame = +1
Query: 28 APFNNQQLVGLPQSGQPAMIQNQLLKQIPAVSGSASLLPLQQPQSQQQLASSAQLQQSSL 87
A +NQ L L + Q Q QL K + ++ A P Q P Q+A +QQ
Sbjct: 310 AILDNQNLGKLAECAQ---YQAQLQKNLMYLAAIADAQP-QTPALPPQMAPHPAMQQGFY 477
Query: 88 TLNQQQLPQLMQQ-------PKPMGQPQLQQQLQHQQLLQQQLQLQQQYQRLQQQL 136
+ Q QQ P G P Q QHQ QQQ QL QQ + Q L
Sbjct: 478 MQHPQAAAMAQQQGMFPQKMPMQFGNPHQMQDQQHQ---QQQQQLHQQAMQGQMGL 636
Score = 32.7 bits (73), Expect = 0.22
Identities = 45/170 (26%), Positives = 65/170 (37%), Gaps = 11/170 (6%)
Frame = +1
Query: 26 QQAPFNNQQLVGLPQSGQPAMIQNQLLKQIPAVSGSASLLPLQQPQSQQQLASSAQLQQ- 84
QQ P + PQ Q + Q+ K + +L + Q+ +LA AQ Q
Sbjct: 202 QQTPQMIPMMPSFPQ--QTNITTEQIQKYLD--ENKKLILAILDNQNLGKLAECAQYQAQ 369
Query: 85 --------SSLTLNQQQLPQLMQQPKPMGQPQLQQ--QLQHQQLLQQQLQLQQQYQRLQQ 134
+++ Q Q P L Q P P +QQ +QH Q
Sbjct: 370 LQKNLMYLAAIADAQPQTPALPPQMAP--HPAMQQGFYMQHPQ----------------- 492
Query: 135 QLASSAQLQQNSLTLNQQQLPLLMQQPKSMGQPLLQQQQQQLLQQQLQLQ 184
++A QQ + Q++P+ P M QQQQQQL QQ +Q Q
Sbjct: 493 ---AAAMAQQQGMF--PQKMPMQFGNPHQMQDQQHQQQQQQLHQQAMQGQ 627
>TC93379 similar to GP|19569994|gb|AAL92295.1 hypothetical protein
{Dictyostelium discoideum}, partial (1%)
Length = 535
Score = 48.1 bits (113), Expect = 5e-06
Identities = 25/47 (53%), Positives = 29/47 (61%)
Frame = +2
Query: 217 QKSMGQPLLQQQLQQQQLLLQQQQLVLQQQQQQQQQQRTQLLQQQMQ 263
Q+ P QQQ Q QQ QQQ + + QQQQQQQ + QL QQQMQ
Sbjct: 83 QQMQPNPYHQQQQQHQQQQQQQQYMAMMMQQQQQQQMQQQLQQQQMQ 223
Score = 44.3 bits (103), Expect = 7e-05
Identities = 30/93 (32%), Positives = 41/93 (43%), Gaps = 4/93 (4%)
Frame = +2
Query: 195 ASSAQLQQNSSTLNQLPQLMQQQKSMGQPLLQQQLQQQQ----LLLQQQQLVLQQQQQQQ 250
A +A N + Q+ Q QQQ QQQQ ++ QQQQ +QQQ QQQ
Sbjct: 35 AGAAAAAMNGGYYQGMQQMQPNPYHQQQQQHQQQQQQQQYMAMMMQQQQQQQMQQQLQQQ 214
Query: 251 QQQRTQLLQQQMQALRIPGPAGHKSLSLTGSQP 283
Q Q+ + Q + G H S++ G P
Sbjct: 215 QMQQGNMNNMYPQHMMYGGGRPHPSMNYMGPPP 313
Score = 40.0 bits (92), Expect = 0.001
Identities = 25/56 (44%), Positives = 32/56 (56%), Gaps = 4/56 (7%)
Frame = +2
Query: 96 QLMQQPKPMGQPQLQQQLQHQQLLQQQ----LQLQQQYQRLQQQLASSAQLQQNSL 147
Q MQQ +P P QQQ QHQQ QQQ + +QQQ Q+ QQ Q+QQ ++
Sbjct: 74 QGMQQMQP--NPYHQQQQQHQQQQQQQQYMAMMMQQQQQQQMQQQLQQQQMQQGNM 235
Score = 38.9 bits (89), Expect = 0.003
Identities = 23/48 (47%), Positives = 28/48 (57%)
Frame = +2
Query: 141 QLQQNSLTLNQQQLPLLMQQPKSMGQPLLQQQQQQLLQQQLQLQQQYQ 188
Q+Q N QQQ QQ + M ++QQQQQQ +QQQLQ QQ Q
Sbjct: 86 QMQPNPYHQQQQQHQQQQQQQQYMAM-MMQQQQQQQMQQQLQQQQMQQ 226
Score = 35.4 bits (80), Expect = 0.034
Identities = 22/63 (34%), Positives = 29/63 (45%)
Frame = +2
Query: 152 QQLPLLMQQPKSMGQPLLQQQQQQLLQQQLQLQQQYQRLQQQLASSAQLQQNSSTLNQLP 211
Q + + P Q QQQQQQ + +QQQ Q+ QQ Q+QQ + N P
Sbjct: 74 QGMQQMQPNPYHQQQQQHQQQQQQQQYMAMMMQQQQQQQMQQQLQQQQMQQGNMN-NMYP 250
Query: 212 QLM 214
Q M
Sbjct: 251 QHM 259
Score = 34.7 bits (78), Expect = 0.058
Identities = 30/84 (35%), Positives = 40/84 (46%), Gaps = 5/84 (5%)
Frame = +2
Query: 52 LKQIPAVSG-----SASLLPLQQPQSQQQLASSAQLQQSSLTLNQQQLPQLMQQPKPMGQ 106
++ +PAV G +A+ + Q QQ+ + QQ QQQ Q QQ + M
Sbjct: 2 MRSMPAVQGLPAGAAAAAMNGGYYQGMQQMQPNPYHQQ------QQQHQQQQQQQQYMAM 163
Query: 107 PQLQQQLQHQQLLQQQLQLQQQYQ 130
QQQ QQ +QQQLQ QQ Q
Sbjct: 164 MMQQQQ---QQQMQQQLQQQQMQQ 226
Score = 33.1 bits (74), Expect = 0.17
Identities = 26/64 (40%), Positives = 29/64 (44%)
Frame = +2
Query: 169 LQQQQQQLLQQQLQLQQQYQRLQQQLASSAQLQQNSSTLNQLPQLMQQQKSMGQPLLQQQ 228
+QQ Q QQ Q QQ Q+ QQ +A Q QQ Q MQQQ LQQQ
Sbjct: 80 MQQMQPNPYHQQQQQHQQQQQQQQYMAMMMQQQQQ--------QQMQQQ-------LQQQ 214
Query: 229 LQQQ 232
QQ
Sbjct: 215 QMQQ 226
Score = 33.1 bits (74), Expect = 0.17
Identities = 27/83 (32%), Positives = 36/83 (42%)
Frame = +2
Query: 105 GQPQLQQQLQHQQLLQQQLQLQQQYQRLQQQLASSAQLQQNSLTLNQQQLPLLMQQPKSM 164
G Q+Q HQQ QQQ+Q+ QQQ QQ + ++MQQ +
Sbjct: 77 GMQQMQPNPYHQQ--------QQQHQQQQQQ---------------QQYMAMMMQQQQ-- 181
Query: 165 GQPLLQQQQQQLLQQQLQLQQQY 187
Q +QQQ QQ QQ + Y
Sbjct: 182 -QQQMQQQLQQQQMQQGNMNNMY 247
>TC79752 similar to GP|11994670|dbj|BAB02898. gene_id:K7P8.17~unknown
protein {Arabidopsis thaliana}, partial (2%)
Length = 2137
Score = 46.2 bits (108), Expect = 2e-05
Identities = 71/245 (28%), Positives = 93/245 (36%), Gaps = 38/245 (15%)
Frame = +1
Query: 55 IPAVSGSASLLPLQQ-PQSQQQLASSAQLQQSSLTL------------NQQQLPQLMQQP 101
IPA SG +S Q P S QQ AQ QQS L+ NQQQ + +
Sbjct: 577 IPAFSGMSSAFNSQTTPPSVQQYPVHAQ-QQSHLSNPHPHLQGPNHPNNQQQAYIRLAKE 753
Query: 102 KPMGQPQLQQQLQHQQL-----LQQQLQLQQQYQRLQQQLASSAQLQQNSLTLNQQQLPL 156
+ + Q Q Q+ LQ QQL L +Q Q Q Q SS QNS QQ+ +
Sbjct: 754 RQLQQQQQQRYLQQQQLSATNALIPHVQAQAQPPISSPQQNSSQAQPQNS----SQQVSV 921
Query: 157 LMQQPKSMGQPLLQQQQQQ---------------LLQQQLQLQQQYQRLQQQLASSAQLQ 201
P S P+ Q QQQ + Q Q+Q Q Q+Q SA+
Sbjct: 922 SPATPSSPLTPMSSQHQQQKHDLPQPGFSRNPGSSVTSQAVKQRQRQAQQRQYQQSARQH 1101
Query: 202 QNSSTLNQLPQLMQQQKSM-----GQPLLQQQLQQQQLLLQQQQLVLQQQQQQQQQQRTQ 256
N Q Q QQ K + G L+ Q + + Q ++ Q TQ
Sbjct: 1102PNQP---QHAQAQQQAKILKGIGRGNTLIHQNNSVDPSHINGLSVAPGTQPVEKGDQITQ 1272
Query: 257 LLQQQ 261
+ Q Q
Sbjct: 1273MTQGQ 1287
Score = 35.4 bits (80), Expect = 0.034
Identities = 30/103 (29%), Positives = 47/103 (45%), Gaps = 8/103 (7%)
Frame = +1
Query: 170 QQQQQQLLQQQLQLQQQYQRLQ-----QQLASSAQLQQNSSTLNQLPQLMQQQKSMGQPL 224
Q Q+Q++ +L +Q Q ++S+ Q ++ Q P QQQ + P
Sbjct: 508 QGHQRQMMVPELPMQVTQGNSQGIPAFSGMSSAFNSQTTPPSVQQYPVHAQQQSHLSNP- 684
Query: 225 LQQQLQQQQLLLQQQQLVLQ---QQQQQQQQQRTQLLQQQMQA 264
LQ QQQ ++ ++Q QQQQQ+ L QQQ+ A
Sbjct: 685 -HPHLQGPNHPNNQQQAYIRLAKERQLQQQQQQRYLQQQQLSA 810
Score = 32.0 bits (71), Expect = 0.38
Identities = 42/162 (25%), Positives = 60/162 (36%), Gaps = 12/162 (7%)
Frame = +1
Query: 110 QQQLQHQQLLQQQLQLQQQYQRLQQQLASSAQLQQNSLTLNQQQLPLLMQQPKSMGQPLL 169
Q+Q+ +L Q Q Q ++S+ Q ++ Q + Q S P L
Sbjct: 517 QRQMMVPELPMQVTQGNSQGIPAFSGMSSAFNSQTTPPSVQQYPVHAQQQSHLSNPHPHL 696
Query: 170 QQQQQQLLQQQLQLQ--QQYQRLQQQLASSAQLQQNSSTLNQLPQLM----------QQQ 217
Q QQQ ++ ++ Q QQQ Q QQ S+T +P + QQ
Sbjct: 697 QGPNHPNNQQQAYIRLAKERQLQQQQQQRYLQQQQLSATNALIPHVQAQAQPPISSPQQN 876
Query: 218 KSMGQPLLQQQLQQQQLLLQQQQLVLQQQQQQQQQQRTQLLQ 259
S QP Q QQ + L Q QQQ+ L Q
Sbjct: 877 SSQAQP--QNSSQQVSVSPATPSSPLTPMSSQHQQQKHDLPQ 996
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.314 0.125 0.320
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 7,222,509
Number of Sequences: 36976
Number of extensions: 95912
Number of successful extensions: 5364
Number of sequences better than 10.0: 246
Number of HSP's better than 10.0 without gapping: 1381
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 2667
length of query: 358
length of database: 9,014,727
effective HSP length: 97
effective length of query: 261
effective length of database: 5,428,055
effective search space: 1416722355
effective search space used: 1416722355
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)
S2: 59 (27.3 bits)
Medicago: description of AC144645.7


