

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC144617.11 + phase: 0
(688 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
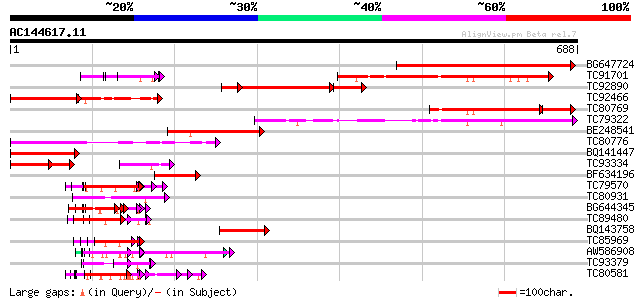
Score E
Sequences producing significant alignments: (bits) Value
BG647724 similar to GP|11141605|gb LEUNIG {Arabidopsis thaliana}... 437 e-123
TC91701 similar to GP|11141605|gb|AAG32022.1 LEUNIG {Arabidopsis... 331 5e-91
TC92890 similar to GP|11141605|gb|AAG32022.1 LEUNIG {Arabidopsis... 233 9e-90
TC92466 similar to GP|11141605|gb|AAG32022.1 LEUNIG {Arabidopsis... 174 2e-73
TC80769 similar to GP|11141605|gb|AAG32022.1 LEUNIG {Arabidopsis... 223 3e-73
TC79322 similar to GP|13605815|gb|AAK32893.1 At2g32700/F24L7.16 ... 244 8e-65
BE248541 similar to GP|11141605|gb| LEUNIG {Arabidopsis thaliana... 201 6e-52
TC80776 similar to GP|13605815|gb|AAK32893.1 At2g32700/F24L7.16 ... 174 1e-43
BQ141447 homologue to GP|11141605|gb| LEUNIG {Arabidopsis thalia... 168 6e-42
TC93334 homologue to GP|13605815|gb|AAK32893.1 At2g32700/F24L7.1... 79 4e-25
BF634196 weakly similar to GP|20804798|dbj putative flower devel... 106 3e-23
TC79570 similar to PIR|F86427|F86427 auxin response factor 6 (AR... 85 1e-16
TC80931 similar to GP|14190783|gb|AAF65166.2 putative phloem tra... 78 1e-14
BG644345 weakly similar to GP|19697333|gb putative protein poten... 75 1e-13
TC89480 similar to GP|20260684|gb|AAM13240.1 unknown protein {Ar... 74 1e-13
BQ143758 weakly similar to PIR|T04859|T048 extensin homolog F28A... 71 1e-12
TC85969 similar to GP|21553908|gb|AAM62991.1 unknown {Arabidopsi... 67 2e-11
AW586908 weakly similar to GP|2104814|emb| pleiotropic effects o... 65 9e-11
TC93379 similar to GP|19569994|gb|AAL92295.1 hypothetical protei... 64 1e-10
TC80581 similar to GP|19683021|gb|AAL92644.1 hypothetical protei... 62 6e-10
>BG647724 similar to GP|11141605|gb LEUNIG {Arabidopsis thaliana}, partial
(25%)
Length = 757
Score = 437 bits (1125), Expect = e-123
Identities = 217/217 (100%), Positives = 217/217 (100%)
Frame = +2
Query: 470 SMSNSFRGNDQVSKSQPGRKRKQPVSSSGPANSSGTANTAGPSPSSAPSTPSGSTHTPGD 529
SMSNSFRGNDQVSKSQPGRKRKQPVSSSGPANSSGTANTAGPSPSSAPSTPSGSTHTPGD
Sbjct: 2 SMSNSFRGNDQVSKSQPGRKRKQPVSSSGPANSSGTANTAGPSPSSAPSTPSGSTHTPGD 181
Query: 530 VISMPSLPHNGSSSKPLMMFSTDGNTSPSNQLADVDRFVADGSLDDNVESFLSHDDTDPR 589
VISMPSLPHNGSSSKPLMMFSTDGNTSPSNQLADVDRFVADGSLDDNVESFLSHDDTDPR
Sbjct: 182 VISMPSLPHNGSSSKPLMMFSTDGNTSPSNQLADVDRFVADGSLDDNVESFLSHDDTDPR 361
Query: 590 DPVGRMDVSKGFTFSELNSVRASTNKVVCSHFSSDGKLLASGGHDKKVVLWYTDSLKQKA 649
DPVGRMDVSKGFTFSELNSVRASTNKVVCSHFSSDGKLLASGGHDKKVVLWYTDSLKQKA
Sbjct: 362 DPVGRMDVSKGFTFSELNSVRASTNKVVCSHFSSDGKLLASGGHDKKVVLWYTDSLKQKA 541
Query: 650 TLEEHSLLITDVRFSPSMPRLATSSYDKTVRVWDVDN 686
TLEEHSLLITDVRFSPSMPRLATSSYDKTVRVWDVDN
Sbjct: 542 TLEEHSLLITDVRFSPSMPRLATSSYDKTVRVWDVDN 652
>TC91701 similar to GP|11141605|gb|AAG32022.1 LEUNIG {Arabidopsis thaliana},
partial (18%)
Length = 1174
Score = 331 bits (849), Expect = 5e-91
Identities = 197/295 (66%), Positives = 216/295 (72%), Gaps = 33/295 (11%)
Frame = +1
Query: 398 GGPPFPRGDPDMLMKLKLAQLQ-------HQQQQNANPQQQQLQQQHSLSNQQSQSANHN 450
GGPPFPRGD DMLMKLKLAQLQ HQQQ + N QQQQLQQ H+LSNQQSQ++NH+
Sbjct: 37 GGPPFPRGDTDMLMKLKLAQLQQQQQQQQHQQQSSINAQQQQLQQ-HALSNQQSQTSNHS 213
Query: 451 MHQQDNKVGGGGGSVTGDGSMSNSFRGNDQVSKSQPGRKRKQPVSSSGPANSSGTANTAG 510
MHQQD KVGGGGGSVT DGSMSNS+RGNDQVSK+Q GRKRKQPVSSSGPANSSGTANTAG
Sbjct: 214 MHQQD-KVGGGGGSVTMDGSMSNSYRGNDQVSKNQMGRKRKQPVSSSGPANSSGTANTAG 390
Query: 511 PSPSSAPSTPSGSTHTPGDVISMPSLPHNGSSSKPLMMFSTDGN---TSPSNQL------ 561
PSPSSAPSTP STHTPGD +SMP+LPHN SSSKPLMMFSTDG TSPSNQL
Sbjct: 391 PSPSSAPSTP--STHTPGDAVSMPALPHNSSSSKPLMMFSTDGTGTLTSPSNQLWDDKDL 564
Query: 562 ---ADVDRFVADGSLDDNVESFLSHDDTDPRDPVGR-MDVSKGFTFSEL----NSVRAST 613
ADVDRFV DGSLDDNVESFLS DDTDPRDPVGR MDVSKG + L N + ++
Sbjct: 565 ELKADVDRFVDDGSLDDNVESFLSQDDTDPRDPVGRCMDVSKGAVNTTLY*S*NFLTSNF 744
Query: 614 NKV---VCSHFSSDGK----LLASGGHD--KKVVLWYTDSLKQKATLEEHSLLIT 659
+V VC FS K L+ D +L YTD + L HS+ +T
Sbjct: 745 QRVPYSVCLIFSRLNKKVCILIFQNLKDICSVTILLYTDIQ*KFIILSHHSIPLT 909
Score = 47.8 bits (112), Expect = 1e-05
Identities = 33/101 (32%), Positives = 43/101 (41%)
Frame = +1
Query: 86 AREQQQQQQQQPQPQQSPHQQQQQQQQHQMQMQQMLLQRQHQQQQQQQQQQQQQQQQQQQ 145
A+ QQQQQQQQ Q Q S + QQQQ QQH + QQ Q + QQ +
Sbjct: 91 AQLQQQQQQQQHQQQSSINAQQQQLQQHALSNQQS--QTSNHSMHQQDKVGGGGGSVTMD 264
Query: 146 QPQHQQQQQQQQQQQQQQGRDRAHLLNGGGANGLVGNPSTA 186
+ Q + Q GR R ++ G G +TA
Sbjct: 265 GSMSNSYRGNDQVSKNQMGRKRKQPVSSSGPANSSGTANTA 387
Score = 46.2 bits (108), Expect = 4e-05
Identities = 30/75 (40%), Positives = 36/75 (48%), Gaps = 7/75 (9%)
Frame = +1
Query: 115 MQMQQMLLQRQHQQQQQQQQQQQQQQQQQQQQPQHQQQQQQQQ-------QQQQQQGRDR 167
M M+ L Q Q QQQQQQ QQQ QQQQ QH QQ Q QQ + G
Sbjct: 70 MLMKLKLAQLQQQQQQQQHQQQSSINAQQQQLQQHALSNQQSQTSNHSMHQQDKVGGGGG 249
Query: 168 AHLLNGGGANGLVGN 182
+ ++G +N GN
Sbjct: 250 SVTMDGSMSNSYRGN 294
Score = 44.7 bits (104), Expect = 1e-04
Identities = 26/63 (41%), Positives = 33/63 (52%), Gaps = 5/63 (7%)
Frame = +1
Query: 131 QQQQQQQQQQQQQQQQPQHQQQQQQQQQQQQQQGRDRAHLLN-----GGGANGLVGNPST 185
Q QQQQQQQQ QQQ QQQQ QQ QQ + H ++ GGG + + S
Sbjct: 94 QLQQQQQQQQHQQQSSINAQQQQLQQHALSNQQSQTSNHSMHQQDKVGGGGGSVTMDGSM 273
Query: 186 ANA 188
+N+
Sbjct: 274 SNS 282
Score = 42.7 bits (99), Expect = 5e-04
Identities = 24/70 (34%), Positives = 34/70 (48%)
Frame = +1
Query: 117 MQQMLLQRQHQQQQQQQQQQQQQQQQQQQQPQHQQQQQQQQQQQQQQGRDRAHLLNGGGA 176
+ ++ L + QQQQQQQ QQQ QQQQ Q QQ Q + + GGG+
Sbjct: 73 LMKLKLAQLQQQQQQQQHQQQSSINAQQQQLQQHALSNQQSQTSNHSMHQQDKVGGGGGS 252
Query: 177 NGLVGNPSTA 186
+ G+ S +
Sbjct: 253 VTMDGSMSNS 282
>TC92890 similar to GP|11141605|gb|AAG32022.1 LEUNIG {Arabidopsis thaliana},
partial (11%)
Length = 547
Score = 233 bits (594), Expect(3) = 9e-90
Identities = 114/117 (97%), Positives = 114/117 (97%)
Frame = +1
Query: 278 PRAAGPEGSLMAVPGSNQGGNNLTLKGWPLTGLEHLRSGLLQQQKPFIQSPQPFHQLPML 337
P GPEGSLMAVPGSNQGGNNLTLKGWPLTGLEHLRSGLLQQQKPFIQSPQPFHQLPML
Sbjct: 79 PELXGPEGSLMAVPGSNQGGNNLTLKGWPLTGLEHLRSGLLQQQKPFIQSPQPFHQLPML 258
Query: 338 TQQHQQQLMLAQQNLASPSASDDSRRLRMLLNPRNMGVSKDGLSNPVGDVVSNVGSP 394
TQQHQQQLMLAQQNLASPSASDDSRRLRMLLNPRNMGVSKDGLSNPVGDVVSNVGSP
Sbjct: 259 TQQHQQQLMLAQQNLASPSASDDSRRLRMLLNPRNMGVSKDGLSNPVGDVVSNVGSP 429
Score = 84.7 bits (208), Expect(3) = 9e-90
Identities = 39/40 (97%), Positives = 39/40 (97%)
Frame = +2
Query: 394 PLQAGGPPFPRGDPDMLMKLKLAQLQHQQQQNANPQQQQL 433
PLQAGGPPFPRGDPDMLMKLKLAQLQHQQQQNANPQQ QL
Sbjct: 428 PLQAGGPPFPRGDPDMLMKLKLAQLQHQQQQNANPQQXQL 547
Score = 53.1 bits (126), Expect(3) = 9e-90
Identities = 25/25 (100%), Positives = 25/25 (100%)
Frame = +3
Query: 258 SQQLPGSTLDIKTEINPVLNPRAAG 282
SQQLPGSTLDIKTEINPVLNPRAAG
Sbjct: 18 SQQLPGSTLDIKTEINPVLNPRAAG 92
>TC92466 similar to GP|11141605|gb|AAG32022.1 LEUNIG {Arabidopsis thaliana},
partial (14%)
Length = 632
Score = 174 bits (441), Expect(2) = 2e-73
Identities = 83/87 (95%), Positives = 85/87 (97%)
Frame = +3
Query: 1 MSQTNWEADKMLDVYIHDYLVKRDLKASAQAFQAEGKVSSDPVAIDAPGGFLFEWWSVFW 60
MSQTNWEADKMLDVYIHDYLVKRDLKASAQAFQAEGKVSSDPVAIDAPGGFLFEWWSVFW
Sbjct: 108 MSQTNWEADKMLDVYIHDYLVKRDLKASAQAFQAEGKVSSDPVAIDAPGGFLFEWWSVFW 287
Query: 61 DIFIARTNEKHSEVAASYIETQLIKAR 87
DIFIARTNEKHSEVAASYIETQL + +
Sbjct: 288 DIFIARTNEKHSEVAASYIETQLTQGK 368
Score = 120 bits (301), Expect(2) = 2e-73
Identities = 71/105 (67%), Positives = 75/105 (70%), Gaps = 3/105 (2%)
Frame = +1
Query: 84 IKAREQQ---QQQQQQPQPQQSPHQQQQQQQQHQMQMQQMLLQRQHQQQQQQQQQQQQQQ 140
+KAREQQ QQQQQQPQPQQS H QQQQQQ MQMQQ+L+QR H QQQQQQQQ QQQ
Sbjct: 358 LKAREQQQQQQQQQQQPQPQQSQHAQQQQQQ--HMQMQQLLMQR-HAQQQQQQQQHQQQP 528
Query: 141 QQQQQQPQHQQQQQQQQQQQQQQGRDRAHLLNGGGANGLVGNPST 185
Q Q QQP Q QQQQ RDR HLLN G ANGL GNP+T
Sbjct: 529 QSQPQQP----------QPQQQQNRDRTHLLN-GSANGLAGNPAT 630
Score = 31.2 bits (69), Expect = 1.4
Identities = 16/41 (39%), Positives = 18/41 (43%)
Frame = +1
Query: 417 QLQHQQQQNANPQQQQLQQQHSLSNQQSQSANHNMHQQDNK 457
Q Q QQQQ PQQ Q QQ + Q Q H Q +
Sbjct: 379 QQQQQQQQQPQPQQSQHAQQQQQQHMQMQQLLMQRHAQQQQ 501
Score = 30.8 bits (68), Expect = 1.8
Identities = 17/44 (38%), Positives = 23/44 (51%)
Frame = +1
Query: 417 QLQHQQQQNANPQQQQLQQQHSLSNQQSQSANHNMHQQDNKVGG 460
Q QHQQQ + PQQ Q QQ QQ++ H ++ N + G
Sbjct: 505 QQQHQQQPQSQPQQPQPQQ------QQNRDRTHLLNGSANGLAG 618
Score = 30.0 bits (66), Expect = 3.1
Identities = 21/54 (38%), Positives = 23/54 (41%), Gaps = 10/54 (18%)
Frame = +1
Query: 410 LMKLKLAQLQH----------QQQQNANPQQQQLQQQHSLSNQQSQSANHNMHQ 453
LMK L L H +QQQ QQQQ Q Q S QQ Q + M Q
Sbjct: 307 LMKSTLRWLHHI*RLN*LKAREQQQQQQQQQQQPQPQQSQHAQQQQQQHMQMQQ 468
Score = 29.6 bits (65), Expect = 4.0
Identities = 20/55 (36%), Positives = 23/55 (41%), Gaps = 4/55 (7%)
Frame = +1
Query: 407 PDMLMKLKLAQLQHQQQQ----NANPQQQQLQQQHSLSNQQSQSANHNMHQQDNK 457
P + Q QH Q Q + QQQQ QQQH QSQ QQ N+
Sbjct: 412 PQQSQHAQQQQQQHMQMQQLLMQRHAQQQQQQQQHQ-QQPQSQPQQPQPQQQQNR 573
Score = 29.3 bits (64), Expect = 5.3
Identities = 20/60 (33%), Positives = 26/60 (43%)
Frame = +1
Query: 409 MLMKLKLAQLQHQQQQNANPQQQQLQQQHSLSNQQSQSANHNMHQQDNKVGGGGGSVTGD 468
M M+ L Q QQQQ QQQQ QQQ QQ Q + + + G + G+
Sbjct: 454 MQMQQLLMQRHAQQQQ----QQQQHQQQPQSQPQQPQPQQQQNRDRTHLLNGSANGLAGN 621
>TC80769 similar to GP|11141605|gb|AAG32022.1 LEUNIG {Arabidopsis thaliana},
partial (40%)
Length = 1362
Score = 223 bits (568), Expect(2) = 3e-73
Identities = 119/154 (77%), Positives = 127/154 (82%), Gaps = 14/154 (9%)
Frame = +2
Query: 510 GPSPSSAPSTPSGSTHTPGDVISMPSLPHNGSSSKPLMMFSTDGN---TSPSNQL----- 561
GPSPSSAPSTPS THTPGD +SMP+LPHN SSSKPLMMFSTDG TSPSNQL
Sbjct: 20 GPSPSSAPSTPS--THTPGDAVSMPALPHNSSSSKPLMMFSTDGTGTLTSPSNQLWDDKD 193
Query: 562 ----ADVDRFVADGSLDDNVESFLSHDDTDPRDPVGR-MDVSKGFTFSELNSVRASTNKV 616
ADVDRFV DGSLDDNVESFLS DDTDPRDPVGR MDVSKGFTFS++NSVRAS++K+
Sbjct: 194 LELKADVDRFVDDGSLDDNVESFLSQDDTDPRDPVGRCMDVSKGFTFSDVNSVRASSSKI 373
Query: 617 VCSHFSSDGKLLASGGHDKKVVLWYTD-SLKQKA 649
C HFSSDGKLLASGGHDKK V+WY LKQKA
Sbjct: 374 ACCHFSSDGKLLASGGHDKKAVIWYRRIHLKQKA 475
Score = 71.2 bits (173), Expect(2) = 3e-73
Identities = 34/40 (85%), Positives = 36/40 (90%)
Frame = +3
Query: 647 QKATLEEHSLLITDVRFSPSMPRLATSSYDKTVRVWDVDN 686
+K LEEHS LITDVRFS SMPRLATSS+DKTVRVWDVDN
Sbjct: 468 KKPILEEHSALITDVRFSASMPRLATSSFDKTVRVWDVDN 587
>TC79322 similar to GP|13605815|gb|AAK32893.1 At2g32700/F24L7.16
{Arabidopsis thaliana}, partial (59%)
Length = 1824
Score = 244 bits (623), Expect = 8e-65
Identities = 171/411 (41%), Positives = 223/411 (53%), Gaps = 20/411 (4%)
Frame = +2
Query: 298 NNLTLKGWPLTGLEHLRSGL-LQQQKPFIQSPQPFHQLPMLTQQHQQQLML---AQQNLA 353
+ LTLKGWPLTG+E +R G Q QKP +QS F LP QQ QQQL+ AQ N+
Sbjct: 2 HQLTLKGWPLTGIEQIRPGFGAQVQKPLLQSANQFQLLP---QQQQQQLLAQVHAQGNIG 172
Query: 354 SPSASDDSRRLRMLLNPRNM-GVSKDGLSNPVGDVVSNVGSPLQAGGPPFPRGDPDMLMK 412
+ D ++P+ + G+++ GL+ ++N GS G P
Sbjct: 173 NSQVYGD-------MDPQRLRGLARGGLNVKDSQPIANDGS---IGSP------------ 286
Query: 413 LKLAQLQHQQQQNANPQQQQLQQQHSLSNQQSQSANHNMHQQDNKVGGGGGSVTGDGSMS 472
Q ++ Q Q Q S+S QQ + Q+N
Sbjct: 287 ---------MQSTSSKQINMPQMQQSISQQQHDPLHSQQLVQNN---------------- 391
Query: 473 NSFRGNDQVSKSQPGRKRKQPVSSSGPANSSGTANTAGPSPSSAPSTPSGSTHTPGDVIS 532
RKRK P +SSG ANS+GT NT GPS +S PSTP S HTPGD ++
Sbjct: 392 ---------------RKRKGP-TSSGAANSTGTGNTLGPS-NSQPSTP--SIHTPGDGVA 514
Query: 533 MP-SLPHNGSSSKPLMMFSTDGN---TSPSNQLAD--VDRFVADGSLDDNVESFLSHDDT 586
M +L + SK LMM+ T+G S +NQL ++ F GSLDDNVESFLS DD
Sbjct: 515 MAGNLQNVAGVSKALMMYGTEGAGGLASSTNQLLQDGMEHFGDVGSLDDNVESFLSQDDG 694
Query: 587 DPRDPVGRM---------DVSKGFTFSELNSVRASTNKVVCSHFSSDGKLLASGGHDKKV 637
D +D G + D SKGF+FSE++S+R S KVVC HFSSDGKLLAS GHDKKV
Sbjct: 695 DGKDLFGTLKRNPAEHATDSSKGFSFSEVSSIRKSNGKVVCCHFSSDGKLLASAGHDKKV 874
Query: 638 VLWYTDSLKQKATLEEHSLLITDVRFSPSMPRLATSSYDKTVRVWDVDNVS 688
VLW ++LK ++T EEH+++ITDVRF P+ +LATSS+D TVR+WD + S
Sbjct: 875 VLWNMETLKTQSTPEEHTVIITDVRFRPNSTQLATSSFDTTVRLWDAADPS 1027
Score = 33.5 bits (75), Expect = 0.28
Identities = 47/207 (22%), Positives = 71/207 (33%), Gaps = 10/207 (4%)
Frame = +2
Query: 116 QMQQMLLQRQHQQQQQQQQQQQQQQQQQQQQPQHQQQQQQQQQQQQQQGRDRAHLLNGGG 175
Q+Q+ LLQ +Q Q QQQQQQ Q H Q Q R L GG
Sbjct: 68 QVQKPLLQSANQFQLLPQQQQQQLLAQ-----VHAQGNIGNSQVYGDMDPQRLRGLARGG 232
Query: 176 AN----------GLVGNPSTANAIATKMYEERLKLPLQRDSLEDAAMKQRFGDQLLDPNH 225
N G +G+P + + +++ +P + S+ QL+ N
Sbjct: 233 LNVKDSQPIANDGSIGSPMQSTS------SKQINMPQMQQSISQQQHDPLHSQQLVQNNR 394
Query: 226 ASILKSSAATGQPSGQVLHGAAGAMSPQVQARSQQLPGSTLDIKTEINPVLNPRAAGPEG 285
K ++G + G + Q S PG + + + V AG
Sbjct: 395 KR--KGPTSSGAANSTGTGNTLGPSNSQPSTPSIHTPGDGVAMAGNLQNV-----AGVSK 553
Query: 286 SLMAVPGSNQGGNNLTLKGWPLTGLEH 312
+LM GG + G+EH
Sbjct: 554 ALMMYGTEGAGGLASSTNQLLQDGMEH 634
Score = 33.1 bits (74), Expect = 0.36
Identities = 25/86 (29%), Positives = 43/86 (49%)
Frame = +2
Query: 597 VSKGFTFSELNSVRASTNKVVCSHFSSDGKLLASGGHDKKVVLWYTDSLKQKATLEEHSL 656
++ G ELNS + C S LL GG+ + + LW+ K T+ H
Sbjct: 1385 LASGDCIHELNSSGNMFHS--CVFHPSYSNLLVIGGY-QSLELWHMAENKCM-TIPAHEG 1552
Query: 657 LITDVRFSPSMPRLATSSYDKTVRVW 682
+I+ + SP +A++S+DK+V++W
Sbjct: 1553 VISALAQSPVTGMVASASHDKSVKIW 1630
Score = 29.6 bits (65), Expect = 4.0
Identities = 14/41 (34%), Positives = 22/41 (53%)
Frame = +2
Query: 102 SPHQQQQQQQQHQMQMQQMLLQRQHQQQQQQQQQQQQQQQQ 142
SP Q +Q + QMQQ + Q+QH QQ Q ++++
Sbjct: 281 SPMQSTSSKQINMPQMQQSISQQQHDPLHSQQLVQNNRKRK 403
>BE248541 similar to GP|11141605|gb| LEUNIG {Arabidopsis thaliana}, partial
(10%)
Length = 376
Score = 201 bits (512), Expect = 6e-52
Identities = 102/121 (84%), Positives = 108/121 (88%), Gaps = 3/121 (2%)
Frame = +3
Query: 192 KMYEERLKLPLQRDSLEDAAMKQRFGD---QLLDPNHASILKSSAATGQPSGQVLHGAAG 248
KMYEERLK P QRDSL+DAAMKQRFG+ QLLDPNHASILKS+AA GQPSGQVLHG AG
Sbjct: 3 KMYEERLKGPPQRDSLDDAAMKQRFGENMGQLLDPNHASILKSAAAGGQPSGQVLHGTAG 182
Query: 249 AMSPQVQARSQQLPGSTLDIKTEINPVLNPRAAGPEGSLMAVPGSNQGGNNLTLKGWPLT 308
MSPQVQARSQQLPGST DIK+EINPVLNPRAAGPEGSL+ + GSNQ NNLTLKGWPLT
Sbjct: 183 GMSPQVQARSQQLPGSTTDIKSEINPVLNPRAAGPEGSLLGISGSNQWNNNLTLKGWPLT 362
Query: 309 G 309
G
Sbjct: 363 G 365
>TC80776 similar to GP|13605815|gb|AAK32893.1 At2g32700/F24L7.16
{Arabidopsis thaliana}, partial (19%)
Length = 1087
Score = 174 bits (441), Expect = 1e-43
Identities = 108/254 (42%), Positives = 134/254 (52%)
Frame = +1
Query: 2 SQTNWEADKMLDVYIHDYLVKRDLKASAQAFQAEGKVSSDPVAIDAPGGFLFEWWSVFWD 61
+Q+NWEADK+LDVYIHDY +KR L +A+AF AEGKVS+DPVAIDAPGGFLFEWWSVFWD
Sbjct: 535 AQSNWEADKVLDVYIHDYFLKRRLHNTAKAFMAEGKVSTDPVAIDAPGGFLFEWWSVFWD 714
Query: 62 IFIARTNEKHSEVAASYIETQLIKAREQQQQQQQQPQPQQSPHQQQQQQQQHQMQMQQML 121
IFI+RTNEKHSE AASYIETQ K REQ Q QQ
Sbjct: 715 IFISRTNEKHSEAAASYIETQQTKVREQPQIQQ--------------------------- 813
Query: 122 LQRQHQQQQQQQQQQQQQQQQQQQQPQHQQQQQQQQQQQQQQGRDRAHLLNGGGANGLVG 181
Q+ QQ+ Q Q++ P H LN + G++G
Sbjct: 814 ----------LQRMQQRNAQLQRRDPNHP---------------GLGGSLNPMNSEGMLG 918
Query: 182 NPSTANAIATKMYEERLKLPLQRDSLEDAAMKQRFGDQLLDPNHASILKSSAATGQPSGQ 241
P A +A KMYE R+K P +D N ++LKS+ + GQ
Sbjct: 919 QP-PAGQLAMKMYEARMKHP-----------------HSIDANRMALLKSATS---HQGQ 1035
Query: 242 VLHGAAGAMSPQVQ 255
++HG +G MS +Q
Sbjct: 1036LVHGNSGNMSAVLQ 1077
>BQ141447 homologue to GP|11141605|gb| LEUNIG {Arabidopsis thaliana}, partial
(9%)
Length = 441
Score = 168 bits (426), Expect = 6e-42
Identities = 81/84 (96%), Positives = 81/84 (96%)
Frame = +2
Query: 1 MSQTNWEADKMLDVYIHDYLVKRDLKASAQAFQAEGKVSSDPVAIDAPGGFLFEWWSVFW 60
MSQTNWEADKMLDVYIHDYLVKRDLKASAQAFQAEGKVSSDPVAIDAPGGFLFEWWSVFW
Sbjct: 188 MSQTNWEADKMLDVYIHDYLVKRDLKASAQAFQAEGKVSSDPVAIDAPGGFLFEWWSVFW 367
Query: 61 DIFIARTNEKHSEVAASYIETQLI 84
DIFIARTN KHSEVAASY TQLI
Sbjct: 368 DIFIARTNXKHSEVAASYXXTQLI 439
>TC93334 homologue to GP|13605815|gb|AAK32893.1 At2g32700/F24L7.16
{Arabidopsis thaliana}, partial (13%)
Length = 727
Score = 79.0 bits (193), Expect(2) = 4e-25
Identities = 37/53 (69%), Positives = 44/53 (82%)
Frame = +3
Query: 1 MSQTNWEADKMLDVYIHDYLVKRDLKASAQAFQAEGKVSSDPVAIDAPGGFLF 53
M+Q+NWEADKMLDVYI+DYLVK+ L +A++F EGKVS DPVAIDAP F F
Sbjct: 243 MAQSNWEADKMLDVYIYDYLVKKKLHNTAKSFMTEGKVSPDPVAIDAPWRFSF 401
Score = 54.7 bits (130), Expect(2) = 4e-25
Identities = 25/32 (78%), Positives = 27/32 (84%), Gaps = 1/32 (3%)
Frame = +1
Query: 48 PGGFLFEWWSVFWDIFIARTN-EKHSEVAASY 78
PGGFLFEWWSVFWDIFIARTN E +AA+Y
Sbjct: 385 PGGFLFEWWSVFWDIFIARTN*ETF*IIAAAY 480
Score = 44.7 bits (104), Expect = 1e-04
Identities = 29/72 (40%), Positives = 41/72 (56%), Gaps = 6/72 (8%)
Frame = +2
Query: 134 QQQQQQQQQQQQQPQHQQQQQQQQQQQQQQGRDRAHL------LNGGGANGLVGNPSTAN 187
Q+ + QQ + ++ Q Q QQ Q +Q Q Q RD H LN + G++G STA
Sbjct: 473 QRILEAQQLKAKEQQLQMQQLQLMRQAQMQRRDPNHPPIGTPPLNAITSEGVLGQ-STAT 649
Query: 188 AIATKMYEERLK 199
A+A KMYE+R+K
Sbjct: 650 ALAAKMYEDRMK 685
>BF634196 weakly similar to GP|20804798|dbj putative flower development
regulator LEUNI protein {Oryza sativa (japonica
cultivar-group)}, partial (4%)
Length = 171
Score = 106 bits (265), Expect = 3e-23
Identities = 53/56 (94%), Positives = 53/56 (94%)
Frame = +3
Query: 176 ANGLVGNPSTANAIATKMYEERLKLPLQRDSLEDAAMKQRFGDQLLDPNHASILKS 231
ANGL GNPSTANAI TKMYEERLKLPLQRDSLEDAAMKQRFG QLLDPNHASILKS
Sbjct: 3 ANGLXGNPSTANAIXTKMYEERLKLPLQRDSLEDAAMKQRFGXQLLDPNHASILKS 170
>TC79570 similar to PIR|F86427|F86427 auxin response factor 6 (ARF6)
[imported] - Arabidopsis thaliana, partial (15%)
Length = 1143
Score = 84.7 bits (208), Expect = 1e-16
Identities = 50/81 (61%), Positives = 52/81 (63%), Gaps = 8/81 (9%)
Frame = +2
Query: 90 QQQQQQQPQPQQS-PHQQQQQ---QQQHQMQMQQMLLQRQH----QQQQQQQQQQQQQQQ 141
Q Q QQ QP Q+ P Q+ Q Q Q Q Q Q LLQ QH Q QQQQQQQQQQ
Sbjct: 425 QNQMLQQSQPHQAFPKNQESQHPSQSQAQTQQFQQLLQHQHSFTNQNYHMQQQQQQQQQQ 604
Query: 142 QQQQQPQHQQQQQQQQQQQQQ 162
QQQQQ QHQQQQQQQ QQQQQ
Sbjct: 605 QQQQQQQHQQQQQQQSQQQQQ 667
Score = 73.6 bits (179), Expect = 2e-13
Identities = 42/91 (46%), Positives = 53/91 (58%), Gaps = 7/91 (7%)
Frame = +2
Query: 76 ASYIETQLIKAREQQQ-----QQQQQPQPQQSPHQQQQQ--QQQHQMQMQQMLLQRQHQQ 128
A+ ++ Q+++ + Q Q+ Q P Q+ QQ QQ Q QH Q +Q+Q QQ
Sbjct: 413 AALMQNQMLQQSQPHQAFPKNQESQHPSQSQAQTQQFQQLLQHQHSFTNQNYHMQQQQQQ 592
Query: 129 QQQQQQQQQQQQQQQQQQPQHQQQQQQQQQQ 159
QQQQQQQQQQQ QQQQQQ QQQQ QQ
Sbjct: 593 QQQQQQQQQQQHQQQQQQQSQQQQQVVDHQQ 685
Score = 64.3 bits (155), Expect = 1e-10
Identities = 46/118 (38%), Positives = 53/118 (43%), Gaps = 7/118 (5%)
Frame = +2
Query: 68 NEKHSEVAASYIETQLIKAREQQQQQQQQPQPQQSPHQQQQQQQQHQMQMQQMLLQRQHQ 127
N++ + S +TQ QQ Q Q Q+ H QQQQQQQ Q Q QQ Q+QHQ
Sbjct: 473 NQESQHPSQSQAQTQQF----QQLLQHQHSFTNQNYHMQQQQQQQQQQQQQQ---QQQHQ 631
Query: 128 QQQQQQQQQQQQQQQQQQQPQHQQQQQQ-------QQQQQQQQGRDRAHLLNGGGANG 178
QQQQQQ QQQQQ QQ Q Q Q Q H N +NG
Sbjct: 632 QQQQQQSQQQQQVVDHQQMSNAVSTMSQFVSAPPSQSTQPMQAISSIGHQQNFSDSNG 805
Score = 55.5 bits (132), Expect = 7e-08
Identities = 37/110 (33%), Positives = 47/110 (42%), Gaps = 11/110 (10%)
Frame = +2
Query: 93 QQQQPQPQQSPHQQQQQQQQHQMQMQQMLLQRQHQQQQQQQQQQQQQQQQQQQQPQH--- 149
Q QQPQ + + Q Q Q + + Q Q Q Q QQ QQ Q QH
Sbjct: 377 QFQQPQNFSNGNAALMQNQMLQQSQPHQAFPKNQESQHPSQSQAQTQQFQQLLQHQHSFT 556
Query: 150 --------QQQQQQQQQQQQQQGRDRAHLLNGGGANGLVGNPSTANAIAT 191
QQQQQQQQQQQQQQ + +V + +NA++T
Sbjct: 557 NQNYHMQQQQQQQQQQQQQQQQQHQQQQQQQSQQQQQVVDHQQMSNAVST 706
Score = 52.8 bits (125), Expect = 4e-07
Identities = 34/77 (44%), Positives = 36/77 (46%), Gaps = 2/77 (2%)
Frame = +2
Query: 89 QQQQQQQQPQPQQSPHQQQQQQQQHQMQMQQMLLQRQHQQQQQQQQQQQQQQQQQQ--QQ 146
QQ Q +Q QQ Q HQ + Q Q Q Q QQ QQ Q Q Q
Sbjct: 383 QQPQNFSNGNAALMQNQMLQQSQPHQAFPKNQESQHPSQSQAQTQQFQQLLQHQHSFTNQ 562
Query: 147 PQHQQQQQQQQQQQQQQ 163
H QQQQQQQQQQQQQ
Sbjct: 563 NYHMQQQQQQQQQQQQQ 613
Score = 36.6 bits (83), Expect = 0.033
Identities = 39/133 (29%), Positives = 53/133 (39%), Gaps = 7/133 (5%)
Frame = +2
Query: 417 QLQHQQQQNANPQQQQLQQQHSLSNQQSQSANHNMHQQDNKVGGGGGSVTGDGSMSN--- 473
Q Q QQQQ QQQ QQQ S QQ Q +H Q N V V+ S S
Sbjct: 581 QQQQQQQQQQQQQQQHQQQQQQQSQQQQQVVDH--QQMSNAVSTMSQFVSAPPSQSTQPM 754
Query: 474 ---SFRGNDQVSKSQPGRKRKQPVSSSGPANSSGTANTAG-PSPSSAPSTPSGSTHTPGD 529
S G+ Q G + G ++ T++ P P+S S ST P
Sbjct: 755 QAISSIGHQQNFSDSNGNPVSPLHNMLGSFSNDETSHLLNFPRPNSWVPVQS-STARPSK 931
Query: 530 VISMPSLPHNGSS 542
+++ L +G+S
Sbjct: 932 RVAVDPLLSSGAS 970
Score = 35.4 bits (80), Expect = 0.073
Identities = 28/99 (28%), Positives = 38/99 (38%), Gaps = 2/99 (2%)
Frame = +2
Query: 417 QLQHQQQQNANPQQQQLQQQHSLSNQQSQSANHNMHQQDNKVGGGGGSVTGDGSMSNSFR 476
Q H QQQ QQQQ QQQ QQ Q + D++ MSN+
Sbjct: 560 QNYHMQQQQQQQQQQQQQQQQQHQQQQQQQSQQQQQVVDHQ------------QMSNAVS 703
Query: 477 GNDQVSKSQPGRKRK--QPVSSSGPANSSGTANTAGPSP 513
Q + P + + Q +SS G + +N SP
Sbjct: 704 TMSQFVSAPPSQSTQPMQAISSIGHQQNFSDSNGNPVSP 820
Score = 32.3 bits (72), Expect = 0.62
Identities = 20/77 (25%), Positives = 29/77 (36%)
Frame = +2
Query: 67 TNEKHSEVAASYIETQLIKAREQQQQQQQQPQPQQSPHQQQQQQQQHQMQMQQMLLQRQH 126
TN+ + + Q + ++QQ QQQQQ Q QQ QQ + + +
Sbjct: 554 TNQNYHMQQQQQQQQQQQQQQQQQHQQQQQQQSQQQQQVVDHQQMSNAVSTMSQFVSAPP 733
Query: 127 QQQQQQQQQQQQQQQQQ 143
Q Q Q QQ
Sbjct: 734 SQSTQPMQAISSIGHQQ 784
>TC80931 similar to GP|14190783|gb|AAF65166.2 putative phloem transcription
factor M1 {Apium graveolens}, partial (67%)
Length = 1193
Score = 78.2 bits (191), Expect = 1e-14
Identities = 53/120 (44%), Positives = 66/120 (54%), Gaps = 3/120 (2%)
Frame = +2
Query: 77 SYIETQLIKAREQQQQQQQQPQPQQSPHQQQQQQQQHQMQMQQMLLQRQHQQQQQQQQQQ 136
S I QL + +QQQQQQQ PQ QQ+ Q QQQ Q+Q Q+Q Q Q QQQQ
Sbjct: 548 SSIFPQLQQQMQQQQQQQQLPQLQQNQQLSQIQQQIPQLQ------QQQQQLPQLQQQQL 709
Query: 137 QQQQQQQQQQPQHQQQQQQQQQQQQQQ---GRDRAHLLNGGGANGLVGNPSTANAIATKM 193
Q QQQQQQ PQ QQ Q QQ QQQQ G + ++ GG + +V ++ AT +
Sbjct: 710 SQLQQQQQQLPQLQQLQHQQLPQQQQMVGAGMGQTYVQGPGGRSQMVSQGQVSSQGATNI 889
Score = 40.8 bits (94), Expect = 0.002
Identities = 25/62 (40%), Positives = 29/62 (46%)
Frame = +1
Query: 131 QQQQQQQQQQQQQQQQPQHQQQQQQQQQQQQQQGRDRAHLLNGGGANGLVGNPSTANAIA 190
Q QQQQQQ QQQQ QHQQ Q QQQ HL + +TA+ A
Sbjct: 472 QLSGQQQQQQMQQQQMQQHQQMQSQQQ-----------HLSTIAATDATTTTTATASTAA 618
Query: 191 TK 192
T+
Sbjct: 619 TE 624
Score = 39.7 bits (91), Expect = 0.004
Identities = 36/96 (37%), Positives = 41/96 (42%), Gaps = 5/96 (5%)
Frame = +2
Query: 414 KLAQLQHQQQQNANPQQQ---QLQQQHSLSNQQSQSANHNMHQQDNKVGGGGGS--VTGD 468
++ QLQ QQQQ QQQ QLQQQ Q Q + + QQ VG G G V G
Sbjct: 650 QIPQLQQQQQQLPQLQQQQLSQLQQQQQQLPQLQQLQHQQLPQQQQMVGAGMGQTYVQGP 829
Query: 469 GSMSNSFRGNDQVSKSQPGRKRKQPVSSSGPANSSG 504
G S VS+ Q VSS G N G
Sbjct: 830 GGRS------QMVSQGQ--------VSSQGATNIGG 895
Score = 38.5 bits (88), Expect = 0.009
Identities = 19/33 (57%), Positives = 22/33 (66%)
Frame = +1
Query: 120 MLLQRQHQQQQQQQQQQQQQQQQQQQQPQHQQQ 152
M++ + QQQQQQ QQQQ QQ QQ Q QQQ
Sbjct: 454 MVVFKPQLSGQQQQQQMQQQQMQQHQQMQSQQQ 552
Score = 38.1 bits (87), Expect = 0.011
Identities = 17/23 (73%), Positives = 17/23 (73%)
Frame = +1
Query: 127 QQQQQQQQQQQQQQQQQQQQPQH 149
QQQQQ QQQQ QQ QQ Q Q QH
Sbjct: 487 QQQQQMQQQQMQQHQQMQSQQQH 555
Score = 38.1 bits (87), Expect = 0.011
Identities = 21/37 (56%), Positives = 22/37 (58%)
Frame = +1
Query: 98 QPQQSPHQQQQQQQQHQMQMQQMLLQRQHQQQQQQQQ 134
+PQ S QQQQQ QQ QMQ QHQQ Q QQQ
Sbjct: 466 KPQLSGQQQQQQMQQQQMQ--------QHQQMQSQQQ 552
Score = 36.6 bits (83), Expect = 0.033
Identities = 16/24 (66%), Positives = 16/24 (66%)
Frame = +1
Query: 90 QQQQQQQPQPQQSPHQQQQQQQQH 113
QQQQQQ Q Q HQQ Q QQQH
Sbjct: 484 QQQQQQMQQQQMQQHQQMQSQQQH 555
Score = 36.2 bits (82), Expect = 0.043
Identities = 17/27 (62%), Positives = 19/27 (69%)
Frame = +1
Query: 119 QMLLQRQHQQQQQQQQQQQQQQQQQQQ 145
Q+ Q+Q QQ QQQQ QQ QQ Q QQQ
Sbjct: 472 QLSGQQQQQQMQQQQMQQHQQMQSQQQ 552
Score = 34.3 bits (77), Expect = 0.16
Identities = 25/64 (39%), Positives = 32/64 (49%)
Frame = +2
Query: 410 LMKLKLAQLQHQQQQNANPQQQQLQQQHSLSNQQSQSANHNMHQQDNKVGGGGGSVTGDG 469
L + +L+QLQ QQQQ PQ QQL QH QQ Q M Q + GG + G
Sbjct: 692 LQQQQLSQLQQQQQQ--LPQLQQL--QHQQLPQQQQMVGAGMGQTYVQGPGGRSQMVSQG 859
Query: 470 SMSN 473
+S+
Sbjct: 860 QVSS 871
Score = 32.3 bits (72), Expect = 0.62
Identities = 28/103 (27%), Positives = 45/103 (43%), Gaps = 2/103 (1%)
Frame = +1
Query: 423 QQNANPQQQQLQQQHSLSNQQSQSANHNMHQQDNKVGGGGGSVTGDGSMSNSFRGNDQVS 482
Q + QQQQ+QQQ +QQ QS ++ + T + + S D +
Sbjct: 472 QLSGQQQQQQMQQQQMQQHQQMQSQQQHLSTIAATDATTTTTATASTAATESAAFPDSTA 651
Query: 483 KS-QPGRKRKQPVSSSGPA-NSSGTANTAGPSPSSAPSTPSGS 523
S P P +S+ A +++ TA TA ++A ST S +
Sbjct: 652 DSTTPATAAATPSTSTAAAFSAAATAATASAVTATATSTASSA 780
Score = 30.4 bits (67), Expect = 2.4
Identities = 16/29 (55%), Positives = 16/29 (55%)
Frame = +1
Query: 94 QQQPQPQQSPHQQQQQQQQHQMQMQQMLL 122
Q Q QQ QQQQ QQ QMQ QQ L
Sbjct: 472 QLSGQQQQQQMQQQQMQQHQQMQSQQQHL 558
Score = 28.5 bits (62), Expect = 9.0
Identities = 18/39 (46%), Positives = 18/39 (46%), Gaps = 1/39 (2%)
Frame = +2
Query: 417 QLQHQQQQNANPQQ-QQLQQQHSLSNQQSQSANHNMHQQ 454
QLQ Q QQ QQ QLQQ LS Q Q QQ
Sbjct: 563 QLQQQMQQQQQQQQLPQLQQNQQLSQIQQQIPQLQQQQQ 679
>BG644345 weakly similar to GP|19697333|gb putative protein potential
transcriptional repressor Not4hp - Mus musculus, partial
(7%)
Length = 635
Score = 74.7 bits (182), Expect = 1e-13
Identities = 46/82 (56%), Positives = 48/82 (58%), Gaps = 11/82 (13%)
Frame = +1
Query: 90 QQQQQQQPQPQQSPHQQQ-----------QQQQQHQMQMQQMLLQRQHQQQQQQQQQQQQ 138
Q QQQ P Q+P QQQ QQQQQ Q Q QQ Q+Q QQQQQQQ QQQQ
Sbjct: 229 QTPGQQQASPFQTPGQQQPSPFSQITPFSQQQQQLQFQQQQQQQQQQLQQQQQQQLQQQQ 408
Query: 139 QQQQQQQQPQHQQQQQQQQQQQ 160
Q Q QQQQ Q QQ Q QQQQ
Sbjct: 409 QLQLQQQQQQLLQQHQSSQQQQ 474
Score = 72.4 bits (176), Expect = 5e-13
Identities = 45/81 (55%), Positives = 47/81 (57%), Gaps = 7/81 (8%)
Frame = +1
Query: 90 QQQQQQQPQPQQSPHQQQQ-------QQQQHQMQMQQMLLQRQHQQQQQQQQQQQQQQQQ 142
Q QQQ P Q+P QQQ QQQ Q+Q Q Q QQQQQQQQQQ Q
Sbjct: 196 QTPGQQQSSPFQTPGQQQASPFQTPGQQQPSPFSQITPFSQQQQQLQFQQQQQQQQQQLQ 375
Query: 143 QQQQPQHQQQQQQQQQQQQQQ 163
QQQQ Q QQQQQ Q QQQQQQ
Sbjct: 376 QQQQQQLQQQQQLQLQQQQQQ 438
Score = 68.6 bits (166), Expect = 8e-12
Identities = 46/94 (48%), Positives = 49/94 (51%), Gaps = 12/94 (12%)
Frame = +1
Query: 90 QQQQQQQPQPQQSPHQQQQQQQQHQMQMQQMLLQRQHQQQ------------QQQQQQQQ 137
Q QQQ P Q+P QQQ Q Q Q Q QQQ QQQQ Q Q
Sbjct: 163 QTPGQQQASPFQTPGQQQSSPFQTPGQQQASPFQTPGQQQPSPFSQITPFSQQQQQLQFQ 342
Query: 138 QQQQQQQQQPQHQQQQQQQQQQQQQQGRDRAHLL 171
QQQQQQQQQ Q QQQQQ QQQQQ Q + + LL
Sbjct: 343 QQQQQQQQQLQQQQQQQLQQQQQLQLQQQQQQLL 444
Score = 60.5 bits (145), Expect = 2e-09
Identities = 35/56 (62%), Positives = 36/56 (63%)
Frame = +1
Query: 89 QQQQQQQQPQPQQSPHQQQQQQQQHQMQMQQMLLQRQHQQQQQQQQQQQQQQQQQQ 144
QQQQQ Q Q QQ QQ QQQQQ Q+Q QQ L Q QQQQQQ QQ Q QQQQ
Sbjct: 316 QQQQQLQFQQQQQQQQQQLQQQQQQQLQQQQQL---QLQQQQQQLLQQHQSSQQQQ 474
Score = 60.1 bits (144), Expect = 3e-09
Identities = 32/60 (53%), Positives = 37/60 (61%)
Frame = +1
Query: 81 TQLIKAREQQQQQQQQPQPQQSPHQQQQQQQQHQMQMQQMLLQRQHQQQQQQQQQQQQQQ 140
+Q+ +QQQQ Q Q Q QQ Q QQQQQQ Q QQ+ LQ+Q QQ QQ Q QQQQ
Sbjct: 295 SQITPFSQQQQQLQFQQQQQQQQQQLQQQQQQQLQQQQQLQLQQQQQQLLQQHQSSQQQQ 474
Score = 57.8 bits (138), Expect = 1e-08
Identities = 32/62 (51%), Positives = 41/62 (65%)
Frame = +1
Query: 72 SEVAASYIETQLIKAREQQQQQQQQPQPQQSPHQQQQQQQQHQMQMQQMLLQRQHQQQQQ 131
S++ + Q ++ ++QQQQQQQQ Q QQ QQ QQQQQ Q+Q QQ L +QHQ QQ
Sbjct: 295 SQITPFSQQQQQLQFQQQQQQQQQQLQQQQ--QQQLQQQQQLQLQQQQQQLLQQHQSSQQ 468
Query: 132 QQ 133
QQ
Sbjct: 469 QQ 474
Score = 48.5 bits (114), Expect = 8e-06
Identities = 36/76 (47%), Positives = 37/76 (48%), Gaps = 5/76 (6%)
Frame = +1
Query: 93 QQQQPQPQQSPHQQQQQQQQHQMQMQQMLLQRQHQQQQQQQQQQQQQQQQQQQQ--PQHQ 150
Q QQP Q+P QQQ Q Q Q Q QQQ Q QQQ Q P Q
Sbjct: 139 QAQQPSLFQTPGQQQASPFQTPGQQQSSPFQTPGQQQASPFQTPGQQQPSPFSQITPFSQ 318
Query: 151 QQQQ---QQQQQQQQQ 163
QQQQ QQQQQQQQQ
Sbjct: 319 QQQQLQFQQQQQQQQQ 366
Score = 40.0 bits (92), Expect = 0.003
Identities = 21/42 (50%), Positives = 25/42 (59%)
Frame = +1
Query: 80 ETQLIKAREQQQQQQQQPQPQQSPHQQQQQQQQHQMQMQQML 121
+ Q + ++QQQQQ QQ Q Q QQQQ QQHQ QQ L
Sbjct: 352 QQQQQQLQQQQQQQLQQQQQLQLQQQQQQLLQQHQSSQQQQL 477
Score = 38.5 bits (88), Expect = 0.009
Identities = 30/84 (35%), Positives = 35/84 (40%), Gaps = 7/84 (8%)
Frame = +1
Query: 381 SNPVGDVVSNVGSPLQAGGP----PFPRGDPDMLMKLKLA---QLQHQQQQNANPQQQQL 433
S+P SP Q G PF + P + +L Q Q QQQQ QQQQL
Sbjct: 217 SSPFQTPGQQQASPFQTPGQQQPSPFSQITPFSQQQQQLQFQQQQQQQQQQLQQQQQQQL 396
Query: 434 QQQHSLSNQQSQSANHNMHQQDNK 457
QQQ L QQ Q HQ +
Sbjct: 397 QQQQQLQLQQQQQQLLQQHQSSQQ 468
Score = 34.7 bits (78), Expect = 0.13
Identities = 18/34 (52%), Positives = 20/34 (57%)
Frame = +1
Query: 410 LMKLKLAQLQHQQQQNANPQQQQLQQQHSLSNQQ 443
L + + QLQ QQQ QQQQL QQH S QQ
Sbjct: 370 LQQQQQQQLQQQQQLQLQQQQQQLLQQHQSSQQQ 471
>TC89480 similar to GP|20260684|gb|AAM13240.1 unknown protein {Arabidopsis
thaliana}, partial (43%)
Length = 1068
Score = 74.3 bits (181), Expect = 1e-13
Identities = 50/90 (55%), Positives = 53/90 (58%), Gaps = 7/90 (7%)
Frame = +1
Query: 90 QQQQQQQPQPQQSPHQQQQQQQQ--HQMQMQQMLLQRQHQQQQQQQQQQQQQQQQQQQQP 147
Q Q Q Q Q Q P+ QQQQQ HQ Q QQ Q Q Q Q QQQQQQQ QQQQQQ
Sbjct: 112 QIQSQSQSQSNQDPNLYLQQQQQFLHQQQ-QQPFQQSQTTQSQFQQQQQQQLYQQQQQQR 288
Query: 148 QHQQQQQQQQQQQQQQGRDRA-----HLLN 172
QQQQQQ QQ QQQQ ++ HLLN
Sbjct: 289 ILQQQQQQLQQPQQQQNLHQSLASHFHLLN 378
Score = 63.9 bits (154), Expect = 2e-10
Identities = 34/63 (53%), Positives = 40/63 (62%)
Frame = +1
Query: 78 YIETQLIKAREQQQQQQQQPQPQQSPHQQQQQQQQHQMQMQQMLLQRQHQQQQQQQQQQQ 137
Y++ Q +QQQQ QQ Q QS QQQQQQQ +Q Q QQ +LQ+Q QQ QQ QQQQ
Sbjct: 160 YLQQQQQFLHQQQQQPFQQSQTTQSQFQQQQQQQLYQQQQQQRILQQQQQQLQQPQQQQN 339
Query: 138 QQQ 140
Q
Sbjct: 340 LHQ 348
Score = 63.9 bits (154), Expect = 2e-10
Identities = 39/84 (46%), Positives = 46/84 (54%), Gaps = 10/84 (11%)
Frame = +1
Query: 97 PQPQQSPHQQQQQQQQHQ------MQMQQMLLQRQHQQQQQQQQQQQQQQQQQQQQPQHQ 150
P P +P Q Q Q Q +Q QQ L +Q QQ QQ Q Q Q QQQQQQ +Q
Sbjct: 91 PTPNSNPQIQSQSQSQSNQDPNLYLQQQQQFLHQQQQQPFQQSQTTQSQFQQQQQQQLYQ 270
Query: 151 QQQQ----QQQQQQQQQGRDRAHL 170
QQQQ QQQQQQ QQ + + +L
Sbjct: 271 QQQQQRILQQQQQQLQQPQQQQNL 342
Score = 42.4 bits (98), Expect = 6e-04
Identities = 30/79 (37%), Positives = 38/79 (47%)
Frame = +1
Query: 71 HSEVAASYIETQLIKAREQQQQQQQQPQPQQSPHQQQQQQQQHQMQMQQMLLQRQHQQQQ 130
H + + ++Q +++ QQQQQQQ +QQQQQ QR QQQQ
Sbjct: 187 HQQQQQPFQQSQTTQSQFQQQQQQQL-------YQQQQQ-------------QRILQQQQ 306
Query: 131 QQQQQQQQQQQQQQQQPQH 149
QQ QQ QQQQ Q H
Sbjct: 307 QQLQQPQQQQNLHQSLASH 363
Score = 34.3 bits (77), Expect = 0.16
Identities = 18/47 (38%), Positives = 24/47 (50%)
Frame = +1
Query: 80 ETQLIKAREQQQQQQQQPQPQQSPHQQQQQQQQHQMQMQQMLLQRQH 126
++Q + ++QQ QQQQ Q QQQ QQ Q Q + Q L H
Sbjct: 229 QSQFQQQQQQQLYQQQQQQRILQQQQQQLQQPQQQQNLHQSLASHFH 369
Score = 32.0 bits (71), Expect = 0.81
Identities = 20/46 (43%), Positives = 25/46 (53%), Gaps = 1/46 (2%)
Frame = +1
Query: 70 KHSEVAASYIETQLIKAREQQQQQQQQPQPQQSPHQQQQQQQQ-HQ 114
+ S+ S + Q + QQQQQQ+ Q QQ QQ QQQQ HQ
Sbjct: 211 QQSQTTQSQFQQQQQQQLYQQQQQQRILQQQQQQLQQPQQQQNLHQ 348
Score = 30.8 bits (68), Expect = 1.8
Identities = 20/76 (26%), Positives = 31/76 (40%)
Frame = +1
Query: 80 ETQLIKAREQQQQQQQQPQPQQSPHQQQQQQQQHQMQMQQMLLQRQHQQQQQQQQQQQQQ 139
+ QL + ++QQ+ QQQ Q Q P QQQ Q + L + + QQ
Sbjct: 253 QQQLYQQQQQQRILQQQQQQLQQPQQQQNLHQSLASHFHLLNLVENLAEVVEHGNPDQQS 432
Query: 140 QQQQQQQPQHQQQQQQ 155
+ H ++ QQ
Sbjct: 433 DALITELSNHFEKCQQ 480
Score = 28.5 bits (62), Expect = 9.0
Identities = 14/31 (45%), Positives = 16/31 (51%)
Frame = +1
Query: 419 QHQQQQNANPQQQQLQQQHSLSNQQSQSANH 449
Q QQQ+ QQQQLQQ N A+H
Sbjct: 271 QQQQQRILQQQQQQLQQPQQQQNLHQSLASH 363
Score = 28.5 bits (62), Expect = 9.0
Identities = 19/41 (46%), Positives = 20/41 (48%), Gaps = 4/41 (9%)
Frame = +1
Query: 417 QLQHQQQQNANPQQQQLQQQHSLSNQQSQ----SANHNMHQ 453
Q Q QQQQ QQQ QQQ L QQ Q N+HQ
Sbjct: 229 QSQFQQQQQQQLYQQQ-QQQRILQQQQQQLQQPQQQQNLHQ 348
Score = 28.5 bits (62), Expect = 9.0
Identities = 16/43 (37%), Positives = 19/43 (43%)
Frame = +1
Query: 121 LLQRQHQQQQQQQQQQQQQQQQQQQQPQHQQQQQQQQQQQQQQ 163
++ + Q Q Q Q Q Q Q QQQ QQQQQ Q
Sbjct: 85 MIPTPNSNPQIQSQSQSQSNQDPNLYLQQQQQFLHQQQQQPFQ 213
>BQ143758 weakly similar to PIR|T04859|T048 extensin homolog F28A21.80 -
Arabidopsis thaliana, partial (5%)
Length = 856
Score = 71.2 bits (173), Expect = 1e-12
Identities = 42/62 (67%), Positives = 45/62 (71%), Gaps = 1/62 (1%)
Frame = +1
Query: 255 QARSQQLPGSTLDIKTEINPVLNPRAAGPEGSLMAVPGSN-QGGNNLTLKGWPLTGLEHL 313
QARSQ LPGSTLDIKTEINPVLN +AAGPE S MAV + + LKGWPLTGLE
Sbjct: 10 QARSQ*LPGSTLDIKTEINPVLNSKAAGPERSSMAVFWIKLRRFHY*PLKGWPLTGLEQS 189
Query: 314 RS 315
S
Sbjct: 190 TS 195
>TC85969 similar to GP|21553908|gb|AAM62991.1 unknown {Arabidopsis thaliana},
partial (31%)
Length = 2756
Score = 67.0 bits (162), Expect = 2e-11
Identities = 37/65 (56%), Positives = 41/65 (62%), Gaps = 4/65 (6%)
Frame = +2
Query: 103 PHQQQQQQQQHQ----MQMQQMLLQRQHQQQQQQQQQQQQQQQQQQQQPQHQQQQQQQQQ 158
PH + Q QQ Q + QQ LQ+Q QQ + QQQQQQ Q Q QQ QPQHQ QQQQQ Q
Sbjct: 1568 PHSRIQVQQHVQDPGYLLQQQFELQQQQQQFELQQQQQQPQHQPQQSQPQHQPQQQQQLQ 1747
Query: 159 QQQQQ 163
QQQ
Sbjct: 1748 HHQQQ 1762
Score = 57.0 bits (136), Expect = 2e-08
Identities = 34/61 (55%), Positives = 36/61 (58%), Gaps = 1/61 (1%)
Frame = +2
Query: 95 QQPQPQQSPHQQQQQQQQHQMQMQQMLLQRQHQQQQQQQQQQQQQQQ-QQQQQPQHQQQQ 153
Q Q Q P QQQ + Q Q QQ LQ+Q QQ Q Q QQ Q Q Q QQQQQ QH QQQ
Sbjct: 1583 QVQQHVQDPGYLLQQQFELQQQQQQFELQQQQQQPQHQPQQSQPQHQPQQQQQLQHHQQQ 1762
Query: 154 Q 154
Q
Sbjct: 1763 Q 1765
Score = 54.7 bits (130), Expect = 1e-07
Identities = 37/95 (38%), Positives = 43/95 (44%), Gaps = 11/95 (11%)
Frame = +2
Query: 78 YIETQLIKAREQQQQQQQQPQPQQSPHQQQQQQQQHQMQMQQMLLQRQHQQQQQ------ 131
Y+ Q + ++QQQQ + Q Q QQ HQ QQ Q QHQ Q QQ L QH QQQQ
Sbjct: 1613 YLLQQQFELQQQQQQFELQQQQQQPQHQPQQSQPQHQPQQQQQL---QHHQQQQFIHGGH 1783
Query: 132 -----QQQQQQQQQQQQQQQPQHQQQQQQQQQQQQ 161
QQQP HQ +Q QQ
Sbjct: 1784 YIHHNPAIPTYYPVYPSQQQPHHQVYYVPARQPQQ 1888
Score = 50.4 bits (119), Expect = 2e-06
Identities = 36/70 (51%), Positives = 40/70 (56%), Gaps = 2/70 (2%)
Frame = +2
Query: 86 AREQQQQQQQQP-QPQQSPHQQQQQQQQHQMQMQQMLLQRQHQQQQQQQQQQ-QQQQQQQ 143
+R Q QQ Q P Q + QQQQQQ ++Q QQ Q QHQ QQ Q Q Q QQQQQ Q
Sbjct: 1574 SRIQVQQHVQDPGYLLQQQFELQQQQQQFELQQQQQ--QPQHQPQQSQPQHQPQQQQQLQ 1747
Query: 144 QQQPQHQQQQ 153
HQQQQ
Sbjct: 1748 ----HHQQQQ 1765
Score = 36.2 bits (82), Expect = 0.043
Identities = 36/114 (31%), Positives = 46/114 (39%), Gaps = 35/114 (30%)
Frame = +2
Query: 85 KAREQQQQQQQQPQPQQ----------SPH-------------QQQQQQQQHQMQMQ--- 118
+A+ Q Q Q Q PQ QQ SP +Q+ Q Q+Q+Q
Sbjct: 1343 QAQSQAQTQSQAPQFQQKSSGGGSDLPSPDSVSSDSSLSNAMSRQKPVVYQEQIQIQPGT 1522
Query: 119 ---------QMLLQRQHQQQQQQQQQQQQQQQQQQQQPQHQQQQQQQQQQQQQQ 163
++ L H + Q QQ Q QQQ QQQQQ + QQQQQQ
Sbjct: 1523 TRVSNPVDPKLNLSDPHSRIQVQQHVQDPGYLLQQQFELQQQQQQFELQQQQQQ 1684
Score = 34.7 bits (78), Expect = 0.13
Identities = 40/135 (29%), Positives = 50/135 (36%), Gaps = 13/135 (9%)
Frame = +2
Query: 340 QHQQQLMLAQQNLASP-SASDDSRRLRMLLNPRNMGVSKDGLSNPVGDVVSNVGSPLQAG 398
Q QQ+ +L SP S S DS + + + + P VSN P
Sbjct: 1382 QFQQKSSGGGSDLPSPDSVSSDSSLSNAMSRQKPVVYQEQIQIQPGTTRVSNPVDPKLNL 1561
Query: 399 GPPFPR-------GDPDMLMKLKLAQLQHQQQQNANPQQQQLQQQHSLSN-----QQSQS 446
P R DP L++ + Q QQQ QQQQ Q Q S QQ Q
Sbjct: 1562 SDPHSRIQVQQHVQDPGYLLQQQFELQQQQQQFELQQQQQQPQHQPQQSQPQHQPQQQQQ 1741
Query: 447 ANHNMHQQDNKVGGG 461
H HQQ + GG
Sbjct: 1742 LQH--HQQQQFIHGG 1780
Score = 28.9 bits (63), Expect = 6.9
Identities = 32/117 (27%), Positives = 45/117 (38%), Gaps = 12/117 (10%)
Frame = +2
Query: 386 DVVSNVGSPLQAGGPPFPRGDPDMLMKLKLAQLQHQQQQNANPQQQQLQQQHSLSNQQSQ 445
D S+ G P+ PP P+ ++ Q+Q Q Q A Q Q QSQ
Sbjct: 1232 DERSDHGVPVGYRKPPTPQP--------QVVQVQPQAQNQAQTQSQA----------QSQ 1357
Query: 446 SANHNMHQQDNKVGGGGG-------SVTGDGSMSNSFRGNDQV-----SKSQPGRKR 490
+ + Q + GGG SV+ D S+SN+ V + QPG R
Sbjct: 1358 AQTQSQAPQFQQKSSGGGSDLPSPDSVSSDSSLSNAMSRQKPVVYQEQIQIQPGTTR 1528
>AW586908 weakly similar to GP|2104814|emb| pleiotropic effects on cellular
differentiation and slug behaviour {Dictyostelium
discoideum}, partial (8%)
Length = 668
Score = 65.1 bits (157), Expect = 9e-11
Identities = 57/192 (29%), Positives = 84/192 (43%), Gaps = 17/192 (8%)
Frame = +2
Query: 98 QPQQSPHQQQQQQQQHQMQMQQMLLQRQHQQQQQQQ--------QQQQQQQQQQQQQPQH 149
QP S QQQQQQQ+ Q + L Q Q QQQ+QQQ Q QQQQ Q QQ PQ
Sbjct: 5 QPPASWSQQQQQQQKMQSLLPMPLDQLQQQQQRQQQLAGSQNLPQPQQQQPQLTQQSPQQ 184
Query: 150 QQQQQQQQQQQQQQGRDRAHLLNGGGANGLVGNPSTANAIATKMY------EERLKLPLQ 203
QQQQQ QQ Q + + + N V P T + + ++ ++ L+ +
Sbjct: 185 PQQQQQHQQPCQNTTMNNGTIGSNQIPNQCVQQPVTYSQLQQQLLSGSMQSQQNLQSAGK 364
Query: 204 RDSLEDAAMKQRFGDQLLDPNHASILKSSAATG---QPSGQVLHGAAGAMSPQVQARSQQ 260
+ + + Q +D A +L+ Q S Q+L + PQ SQ
Sbjct: 365 NGLMMTSLPQDSQFQQQIDQQQAGLLQRQQQQNQLQQSSLQLLQQSMLQRGPQQPQMSQT 544
Query: 261 LPGSTLDIKTEI 272
+P + D + ++
Sbjct: 545 VPQNISDQQQQL 580
Score = 62.8 bits (151), Expect = 4e-10
Identities = 38/75 (50%), Positives = 41/75 (54%)
Frame = +2
Query: 89 QQQQQQQQPQPQQSPHQQQQQQQQHQMQMQQMLLQRQHQQQQQQQQQQQQQQQQQQQQPQ 148
QQQ QQQ Q QQ Q QQ +QQ +LQR QQ Q Q Q QQQQ
Sbjct: 407 QQQIDQQQAGLLQRQQQQNQLQQSSLQLLQQSMLQRGPQQPQMSQTVPQNISDQQQQLQL 586
Query: 149 HQQQQQQQQQQQQQQ 163
Q+ QQQQQQQQQQQ
Sbjct: 587 LQKLQQQQQQQQQQQ 631
Score = 56.6 bits (135), Expect = 3e-08
Identities = 65/213 (30%), Positives = 85/213 (39%), Gaps = 36/213 (16%)
Frame = +2
Query: 89 QQQQQQQQPQ---PQQSPHQQQQQQQQHQMQMQQMLLQRQHQQQQ-QQQQQQQQQQQQQQ 144
QQQQQQQ+ Q P QQQQQ+Q Q+ Q L Q Q QQ Q QQ QQ QQQQQ
Sbjct: 26 QQQQQQQKMQSLLPMPLDQLQQQQQRQQQLAGSQNLPQPQQQQPQLTQQSPQQPQQQQQH 205
Query: 145 QQP---------------------QHQQQQQQQQQQQQQQGRDRAHLLNGGGANGLVGNP 183
QQP Q Q QQQ L G NGL+
Sbjct: 206 QQPCQNTTMNNGTIGSNQIPNQCVQQPVTYSQLQQQLLSGSMQSQQNLQSAGKNGLMMTS 385
Query: 184 STANA---------IATKMYEERLKLPLQRDSLE--DAAMKQRFGDQLLDPNHASILKSS 232
++ A + ++ + LQ+ SL+ +M QR Q P + + +
Sbjct: 386 LPQDSQFQQQIDQQQAGLLQRQQQQNQLQQSSLQLLQQSMLQRGPQQ---PQMSQTVPQN 556
Query: 233 AATGQPSGQVLHGAAGAMSPQVQARSQQLPGST 265
+ Q Q+L + Q Q + QQ P ST
Sbjct: 557 ISDQQQQLQLLQ----KLQQQQQQQQQQQPLST 643
Score = 52.4 bits (124), Expect = 6e-07
Identities = 33/76 (43%), Positives = 38/76 (49%)
Frame = +2
Query: 88 EQQQQQQQQPQPQQSPHQQQQQQQQHQMQMQQMLLQRQHQQQQQQQQQQQQQQQQQQQQP 147
+ Q QQQ Q Q Q+QQQQ Q Q Q+L Q Q+ QQ Q Q Q Q
Sbjct: 392 QDSQFQQQIDQQQAGLLQRQQQQNQLQQSSLQLLQQSMLQRGPQQPQMSQTVPQNISDQQ 571
Query: 148 QHQQQQQQQQQQQQQQ 163
Q Q Q+ QQQQQQQ
Sbjct: 572 QQLQLLQKLQQQQQQQ 619
Score = 47.4 bits (111), Expect = 2e-05
Identities = 44/138 (31%), Positives = 50/138 (35%), Gaps = 53/138 (38%)
Frame = +2
Query: 80 ETQLIKAREQQQQQQQQPQPQQSPHQQQQQQQQHQM------------------------ 115
+ QL ++ Q QQQQPQ Q QQ QQQQQHQ
Sbjct: 104 QQQLAGSQNLPQPQQQQPQLTQQSPQQPQQQQQHQQPCQNTTMNNGTIGSNQIPNQCVQQ 283
Query: 116 -----QMQQMLLQRQHQQQQQ--------------------QQQQQQQQ----QQQQQQQ 146
Q+QQ LL Q QQ QQQ QQQ Q+QQQQ
Sbjct: 284 PVTYSQLQQQLLSGSMQSQQNLQSAGKNGLMMTSLPQDSQFQQQIDQQQAGLLQRQQQQN 463
Query: 147 PQHQQQQQQQQQQQQQQG 164
Q Q QQ Q+G
Sbjct: 464 QLQQSSLQLLQQSMLQRG 517
Score = 45.8 bits (107), Expect = 5e-05
Identities = 33/70 (47%), Positives = 35/70 (49%), Gaps = 11/70 (15%)
Frame = +2
Query: 91 QQQQQQPQPQQSPHQQQQQQQ----QHQMQMQQMLLQRQHQQQQQ-------QQQQQQQQ 139
Q+QQQQ Q QQS Q QQ Q QM Q + Q QQQQ QQQQQQQQ
Sbjct: 443 QRQQQQNQLQQSSLQLLQQSMLQRGPQQPQMSQTVPQNISDQQQQLQLLQKLQQQQQQQQ 622
Query: 140 QQQQQQQPQH 149
QQQ H
Sbjct: 623 QQQPLSTSSH 652
Score = 28.9 bits (63), Expect = 6.9
Identities = 50/193 (25%), Positives = 74/193 (37%), Gaps = 5/193 (2%)
Frame = +2
Query: 253 QVQARSQQLPGS-TLDIKTEINPVLNPRAAGP--EGSLMAVPGSNQGGNNLTLKGWPLTG 309
Q Q R QQL GS L + P L ++ + P N NN T+ G
Sbjct: 89 QQQQRQQQLAGSQNLPQPQQQQPQLTQQSPQQPQQQQQHQQPCQNTTMNNGTI------G 250
Query: 310 LEHLRSGLLQQQKPFIQSPQPFHQLPMLTQQHQQQLMLAQQN-LASPSASDDSRRLRMLL 368
+ + +QQ + Q Q QL + Q QQ L A +N L S DS+ + +
Sbjct: 251 SNQIPNQCVQQPVTYSQLQQ---QLLSGSMQSQQNLQSAGKNGLMMTSLPQDSQFQQQID 421
Query: 369 NPRNMGVSKDGLSNPVGDV-VSNVGSPLQAGGPPFPRGDPDMLMKLKLAQLQHQQQQNAN 427
+ + + N + + + + GP P+ + + Q Q Q Q
Sbjct: 422 QQQAGLLQRQQQQNQLQQSSLQLLQQSMLQRGPQQPQMSQTVPQNISDQQQQLQLLQKLQ 601
Query: 428 PQQQQLQQQHSLS 440
QQQQ QQQ LS
Sbjct: 602 QQQQQQQQQQPLS 640
Score = 28.5 bits (62), Expect = 9.0
Identities = 39/151 (25%), Positives = 51/151 (32%), Gaps = 11/151 (7%)
Frame = +2
Query: 319 QQQKPFIQSPQPFHQLPMLTQQHQQQLMLAQQNLASPSASDDSRRLRMLLNPRNMGVSKD 378
QQQK P P QL QQ +QQ + QNL P + P+ +
Sbjct: 38 QQQKMQSLLPMPLDQLQQ--QQQRQQQLAGSQNLPQPQQQQPQLTQQSPQQPQQQQQHQQ 211
Query: 379 GLSNPV---GDVVSNVGSPLQAGGPPFPRGD-PDMLMKLKLAQLQHQQQQNAN------- 427
N G + SN P Q P L+ + Q+ Q N
Sbjct: 212 PCQNTTMNNGTIGSN-QIPNQCVQQPVTYSQLQQQLLSGSMQSQQNLQSAGKNGLMMTSL 388
Query: 428 PQQQQLQQQHSLSNQQSQSANHNMHQQDNKV 458
PQ Q QQQ Q Q+ QQ N++
Sbjct: 389 PQDSQFQQQ----IDQQQAGLLQRQQQQNQL 469
>TC93379 similar to GP|19569994|gb|AAL92295.1 hypothetical protein
{Dictyostelium discoideum}, partial (1%)
Length = 535
Score = 64.3 bits (155), Expect = 1e-10
Identities = 39/86 (45%), Positives = 41/86 (47%)
Frame = +2
Query: 90 QQQQQQQPQPQQSPHQQQQQQQQHQMQMQQMLLQRQHQQQQQQQQQQQQQQQQQQQQPQH 149
Q QQ QP P HQQQQQ HQQQQQQQQ QQQQQQ
Sbjct: 74 QGMQQMQPNPY---HQQQQQ----------------HQQQQQQQQYMAMMMQQQQQQQMQ 196
Query: 150 QQQQQQQQQQQQQQGRDRAHLLNGGG 175
QQ QQQQ QQ H++ GGG
Sbjct: 197 QQLQQQQMQQGNMNNMYPQHMMYGGG 274
Score = 48.9 bits (115), Expect = 6e-06
Identities = 29/62 (46%), Positives = 30/62 (47%)
Frame = +2
Query: 88 EQQQQQQQQPQPQQSPHQQQQQQQQHQMQMQQMLLQRQHQQQQQQQQQQQQQQQQQQQQP 147
+QQQQ QQQ Q QQ QQQQQ QM QQQ QQQQ QQ P
Sbjct: 110 QQQQQHQQQQQQQQYMAMMMQQQQQQQM-------------QQQLQQQQMQQGNMNNMYP 250
Query: 148 QH 149
QH
Sbjct: 251 QH 256
Score = 43.5 bits (101), Expect = 3e-04
Identities = 26/58 (44%), Positives = 29/58 (49%), Gaps = 6/58 (10%)
Frame = +2
Query: 126 HQQQQQQQQQQQQQQQQQQQQPQHQQQQ------QQQQQQQQQQGRDRAHLLNGGGAN 177
+ Q QQ Q QQQQQ Q Q QQQQ QQQQQQQ QQ + + G N
Sbjct: 68 YYQGMQQMQPNPYHQQQQQHQQQQQQQQYMAMMMQQQQQQQMQQQLQQQQMQQGNMNN 241
Score = 38.1 bits (87), Expect = 0.011
Identities = 24/53 (45%), Positives = 26/53 (48%), Gaps = 8/53 (15%)
Frame = +2
Query: 417 QLQHQQQQNANP-------QQQQLQQQHSLSNQQSQSAN-HNMHQQDNKVGGG 461
Q QHQQQQ QQQQ Q Q L QQ Q N +NM+ Q GGG
Sbjct: 116 QQQHQQQQQQQQYMAMMMQQQQQQQMQQQLQQQQMQQGNMNNMYPQHMMYGGG 274
Score = 35.0 bits (79), Expect = 0.096
Identities = 20/40 (50%), Positives = 22/40 (55%), Gaps = 6/40 (15%)
Frame = +2
Query: 87 REQQQQQQQQPQ------PQQSPHQQQQQQQQHQMQMQQM 120
++Q QQQQQQ Q QQ Q QQQ QQ QMQ M
Sbjct: 116 QQQHQQQQQQQQYMAMMMQQQQQQQMQQQLQQQQMQQGNM 235
Score = 34.3 bits (77), Expect = 0.16
Identities = 21/51 (41%), Positives = 26/51 (50%)
Frame = +2
Query: 78 YIETQLIKAREQQQQQQQQPQPQQSPHQQQQQQQQHQMQMQQMLLQRQHQQ 128
Y + Q ++QQQQQ QQ QQ QQQ Q Q QMQQ + + Q
Sbjct: 104 YHQQQQQHQQQQQQQQYMAMMMQQQQQQQMQQQLQ-QQQMQQGNMNNMYPQ 253
Score = 33.1 bits (74), Expect = 0.36
Identities = 22/68 (32%), Positives = 25/68 (36%), Gaps = 5/68 (7%)
Frame = +2
Query: 87 REQQQQQQQQ-----PQPQQSPHQQQQQQQQHQMQMQQMLLQRQHQQQQQQQQQQQQQQQ 141
+ QQQQQQQQ Q QQ QQQ QQQ Q + QH +
Sbjct: 122 QHQQQQQQQQYMAMMMQQQQQQQMQQQLQQQQMQQGNMNNMYPQHMMYGGGRPHPSMNYM 301
Query: 142 QQQQQPQH 149
P H
Sbjct: 302 GPPPMPSH 325
Score = 28.5 bits (62), Expect = 9.0
Identities = 20/65 (30%), Positives = 29/65 (43%), Gaps = 1/65 (1%)
Frame = +2
Query: 286 SLMAVPGSNQGGNNLTLKGWPLTGLEHLRSGLL-QQQKPFIQSPQPFHQLPMLTQQHQQQ 344
S+ AV G G + G G++ ++ QQQ+ Q Q + M+ QQ QQQ
Sbjct: 8 SMPAVQGLPAGAAAAAMNGGYYQGMQQMQPNPYHQQQQQHQQQQQQQQYMAMMMQQQQQQ 187
Query: 345 LMLAQ 349
M Q
Sbjct: 188 QMQQQ 202
>TC80581 similar to GP|19683021|gb|AAL92644.1 hypothetical protein
{Dictyostelium discoideum}, partial (5%)
Length = 850
Score = 62.4 bits (150), Expect = 6e-10
Identities = 55/154 (35%), Positives = 73/154 (46%), Gaps = 20/154 (12%)
Frame = +1
Query: 75 AASYIETQLIKAREQQQQQQQQPQPQQSPHQQQQQQQQHQMQMQQMLLQRQHQQQQQQQQ 134
+A ++ L ++Q Q QQP+P P QQQ Q Q +Q QQ+ LQ+Q+Q+ QQQ
Sbjct: 217 SAQLQQSSLTLNQQQLPQLMQQPKPMGQPQLQQQLQHQQLLQ-QQLQLQQQYQRLQQQLA 393
Query: 135 QQQQQQQ------QQQ-----QQP--------QHQQQQQQQQQQQQQQGRDRAHLLNGGG 175
Q QQ QQQ QQP Q QQQQ QQQ Q QQ R
Sbjct: 394 SSAQLQQNSLTLNQQQLPLLMQQPKSMGQPLLQQQQQQLLQQQLQLQQQYQRLQQQLASS 573
Query: 176 ANGLVGNPSTANAIATKMYEER-LKLPLQRDSLE 208
A L N ST N + M +++ + PL + L+
Sbjct: 574 AQ-LQQNSSTLNQLPQLMQQQKSMGQPLLQQQLQ 672
Score = 61.6 bits (148), Expect = 1e-09
Identities = 37/84 (44%), Positives = 49/84 (58%)
Frame = +1
Query: 80 ETQLIKAREQQQQQQQQPQPQQSPHQQQQQQQQHQMQMQQMLLQRQHQQQQQQQQQQQQQ 139
+ QL++ + Q QQQ Q+ Q Q + Q QQ Q+ Q++ Q++ Q QQQ QQQ
Sbjct: 499 QQQLLQQQLQLQQQYQRLQQQLASSAQLQQNSSTLNQLPQLMQQQKSMGQPLLQQQLQQQ 678
Query: 140 QQQQQQQPQHQQQQQQQQQQQQQQ 163
Q QQQ QQQQQQQQQQ+ Q
Sbjct: 679 QLLLQQQQLVLQQQQQQQQQQRTQ 750
Score = 59.3 bits (142), Expect = 5e-09
Identities = 50/122 (40%), Positives = 59/122 (47%), Gaps = 37/122 (30%)
Frame = +1
Query: 79 IETQLIKAREQQQQ-----QQQQPQPQQSPHQQQQ---QQQQHQMQMQQMLLQRQHQQQQ 130
++ QL + + QQ QQQ P Q P Q QQQQ Q+ QQ+ LQ+Q+Q+ Q
Sbjct: 376 LQQQLASSAQLQQNSLTLNQQQLPLLMQQPKSMGQPLLQQQQQQLLQQQLQLQQQYQRLQ 555
Query: 131 QQ------------------QQQQQQ--------QQQQQQQQPQHQQQQ---QQQQQQQQ 161
QQ Q QQQ QQQ QQQQ QQQQ QQQQQQQQ
Sbjct: 556 QQLASSAQLQQNSSTLNQLPQLMQQQKSMGQPLLQQQLQQQQLLLQQQQLVLQQQQQQQQ 735
Query: 162 QQ 163
QQ
Sbjct: 736 QQ 741
Score = 53.5 bits (127), Expect = 3e-07
Identities = 30/57 (52%), Positives = 36/57 (62%)
Frame = +1
Query: 91 QQQQQQPQPQQSPHQQQQQQQQHQMQMQQMLLQRQHQQQQQQQQQQQQQQQQQQQQP 147
Q QQQ Q QQQ QQQQ +Q QQ++LQ+Q QQQQQQ+ Q QQQ Q + P
Sbjct: 616 QLMQQQKSMGQPLLQQQLQQQQLLLQQQQLVLQQQQQQQQQQRTQLLQQQMQALRIP 786
Score = 52.0 bits (123), Expect = 8e-07
Identities = 43/123 (34%), Positives = 56/123 (44%)
Frame = +1
Query: 99 PQQSPHQQQQQQQQHQMQMQQMLLQRQHQQQQQQQQQQQQQQQQQQQQPQHQQQQQQQQQ 158
P Q P QQQ Q+Q + L +Q Q Q QQ + Q Q QQQ QHQQ QQQ Q
Sbjct: 178 PLQQPQSQQQLASSAQLQQSSLTLNQQ-QLPQLMQQPKPMGQPQLQQQLQHQQLLQQQLQ 354
Query: 159 QQQQQGRDRAHLLNGGGANGLVGNPSTANAIATKMYEERLKLPLQRDSLEDAAMKQRFGD 218
QQQ R + L + L N T N +++L L +Q+ + Q+
Sbjct: 355 LQQQYQRLQQQLASSA---QLQQNSLTLN-------QQQLPLLMQQPKSMGQPLLQQQQQ 504
Query: 219 QLL 221
QLL
Sbjct: 505 QLL 513
Score = 50.8 bits (120), Expect = 2e-06
Identities = 37/96 (38%), Positives = 46/96 (47%), Gaps = 2/96 (2%)
Frame = +1
Query: 68 NEKHSEVAASYIETQLIKAREQQQQQQ--QQPQPQQSPHQQQQQQQQHQMQMQQMLLQRQ 125
N+ ++ A L+ ++ Q QQQ Q QQS QQQ MQ + + Q Q
Sbjct: 127 NQLLKQIPAVSGSASLLPLQQPQSQQQLASSAQLQQSSLTLNQQQLPQLMQQPKPMGQPQ 306
Query: 126 HQQQQQQQQQQQQQQQQQQQQPQHQQQQQQQQQQQQ 161
QQQ Q QQ QQQ Q QQQ + QQQ Q QQ
Sbjct: 307 LQQQLQHQQLLQQQLQLQQQYQRLQQQLASSAQLQQ 414
Score = 50.4 bits (119), Expect = 2e-06
Identities = 45/156 (28%), Positives = 66/156 (41%), Gaps = 8/156 (5%)
Frame = +1
Query: 91 QQQQQQPQPQQSPHQQQQQQQQHQMQMQQMLLQRQHQQQQQ-QQQQQQQQQQQQQQQPQH 149
QQ Q Q Q S QQ +Q Q+ Q++ Q + Q Q QQQ Q QQ QQQ Q Q
Sbjct: 184 QQPQSQQQLASSAQLQQSSLTLNQQQLPQLMQQPKPMGQPQLQQQLQHQQLLQQQLQLQQ 363
Query: 150 QQQQQQQQQQQQQQGRDRAHLLNGGGANGLVGNP-STANAIATKMYEERLKLPLQRDSLE 208
Q Q+ QQQ Q + + LN L+ P S + + ++ L+ LQ
Sbjct: 364 QYQRLQQQLASSAQLQQNSLTLNQQQLPLLMQQPKSMGQPLLQQQQQQLLQQQLQLQQQY 543
Query: 209 DAAMKQRFGDQLLDPNHAS------ILKSSAATGQP 238
+Q L N ++ +++ + GQP
Sbjct: 544 QRLQQQLASSAQLQQNSSTLNQLPQLMQQQKSMGQP 651
Score = 50.1 bits (118), Expect = 3e-06
Identities = 54/142 (38%), Positives = 63/142 (44%), Gaps = 52/142 (36%)
Frame = +1
Query: 82 QLIKAREQQQQQQQQPQPQQSPHQQQQQ------QQQHQMQMQQM------LLQRQHQQQ 129
QL++ + Q QQQ Q+ Q Q + Q QQ QQQ + MQQ LLQ+Q QQ
Sbjct: 331 QLLQQQLQLQQQYQRLQQQLASSAQLQQNSLTLNQQQLPLLMQQPKSMGQPLLQQQQQQL 510
Query: 130 QQQ----QQQQQQQQQQ---------------------QQQ----QPQHQQQQQQQ---- 156
QQ QQQ Q+ QQQ QQQ QP QQQ QQQ
Sbjct: 511 LQQQLQLQQQYQRLQQQLASSAQLQQNSSTLNQLPQLMQQQKSMGQPLLQQQLQQQQLLL 690
Query: 157 -------QQQQQQQGRDRAHLL 171
QQQQQQQ + R LL
Sbjct: 691 QQQQLVLQQQQQQQQQQRTQLL 756
Score = 35.4 bits (80), Expect = 0.073
Identities = 41/132 (31%), Positives = 52/132 (39%), Gaps = 2/132 (1%)
Frame = +1
Query: 317 LLQQQKPFIQSPQPFHQLPMLTQQHQQQLMLAQQNLASPSASDDSRRLRMLLNPRNMGVS 376
L QQQ P + QP L QQ QQQL+ Q L +RL+ L
Sbjct: 427 LNQQQLPLLMQ-QPKSMGQPLLQQQQQQLLQQQLQL-----QQQYQRLQQQLASSAQLQQ 588
Query: 377 KDGLSNPVGDVVSNVGSPLQAGGPPFPRGDPDMLMKLKLAQLQHQQQQNANPQQQQLQQQ 436
N + ++ S G P + +L+ QL QQQQ QQQQ QQQ
Sbjct: 589 NSSTLNQLPQLMQQQKS----------MGQPLLQQQLQQQQLLLQQQQLVLQQQQQQQQQ 738
Query: 437 H--SLSNQQSQS 446
L QQ Q+
Sbjct: 739 QRTQLLQQQMQA 774
Score = 33.5 bits (75), Expect = 0.28
Identities = 40/132 (30%), Positives = 54/132 (40%), Gaps = 1/132 (0%)
Frame = +1
Query: 317 LLQQQKP-FIQSPQPFHQLPMLTQQHQQQLMLAQQNLASPSASDDSRRLRMLLNPRNMGV 375
L QQQ P +Q P+P Q P L QQ Q Q +L QQ +RL+ L + +
Sbjct: 247 LNQQQLPQLMQQPKPMGQ-PQLQQQLQHQQLLQQQL----QLQQQYQRLQQQL-ASSAQL 408
Query: 376 SKDGLSNPVGDVVSNVGSPLQAGGPPFPRGDPDMLMKLKLAQLQHQQQQNANPQQQQLQQ 435
++ L+ + + P G P L QQQQ QQ QLQQ
Sbjct: 409 QQNSLTLNQQQLPLLMQQPKSMGQP-----------------LLQQQQQQLLQQQLQLQQ 537
Query: 436 QHSLSNQQSQSA 447
Q+ QQ S+
Sbjct: 538 QYQRLQQQLASS 573
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.310 0.126 0.356
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 17,954,938
Number of Sequences: 36976
Number of extensions: 259680
Number of successful extensions: 13184
Number of sequences better than 10.0: 474
Number of HSP's better than 10.0 without gapping: 3220
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 7322
length of query: 688
length of database: 9,014,727
effective HSP length: 103
effective length of query: 585
effective length of database: 5,206,199
effective search space: 3045626415
effective search space used: 3045626415
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.8 bits)
S2: 62 (28.5 bits)
Medicago: description of AC144617.11


