

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC141106.8 - phase: 0
(337 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
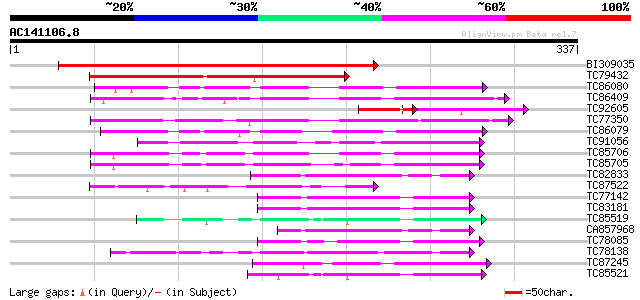
Score E
Sequences producing significant alignments: (bits) Value
BI309035 weakly similar to PIR|C82294|C822 oxidoreductase Tas a... 392 e-109
TC79432 similar to GP|21689703|gb|AAM67473.1 unknown protein {Ar... 150 9e-37
TC86080 similar to PIR|B96632|B96632 hypothetical protein F8A5.2... 87 1e-17
TC86409 similar to PIR|C96632|C96632 hypothetical protein F8A5.2... 84 6e-17
TC92605 similar to GP|21689703|gb|AAM67473.1 unknown protein {Ar... 58 1e-15
TC77350 similar to PIR|T07394|T07394 probable potassium channel ... 80 1e-15
TC86079 similar to GP|15623896|dbj|BAB67953. putative auxin-indu... 79 2e-15
TC91056 similar to GP|16555790|emb|CAD10386. L-galactose dehydro... 67 1e-11
TC85706 similar to GP|21700895|gb|AAM70571.1 At1g60730/F8A5_24 {... 62 2e-10
TC85705 similar to PIR|T12582|T12582 auxin-induced protein - com... 61 5e-10
TC82833 similar to PIR|T48188|T48188 aldose reductase-like prote... 55 5e-08
TC87522 similar to GP|15215594|gb|AAK91342.1 At1g06690/F4H5_17 {... 52 4e-07
TC77142 homologue to GP|9927482|emb|CAC04887.1 unnamed protein p... 51 7e-07
TC83181 similar to PIR|T02543|T02543 aldehyde dehydrogenase homo... 49 2e-06
TC85519 homologue to GP|537298|gb|AAB41556.1|| chalcone reductas... 49 3e-06
CA857968 similar to PIR|T48188|T48 aldose reductase-like protein... 48 6e-06
TC78085 similar to GP|9927482|emb|CAC04887.1 unnamed protein pro... 48 6e-06
TC78138 similar to GP|2792155|emb|CAA11226.1 chalcone reductase ... 47 1e-05
TC87245 similar to GP|10334991|gb|AAG15839.2 NADPH-dependent man... 46 2e-05
TC85521 homologue to GP|537298|gb|AAB41556.1|| chalcone reductas... 45 4e-05
>BI309035 weakly similar to PIR|C82294|C822 oxidoreductase Tas aldo/keto
reductase family VC0667 [imported] - Vibrio cholerae,
partial (27%)
Length = 570
Score = 392 bits (1007), Expect = e-109
Identities = 190/190 (100%), Positives = 190/190 (100%)
Frame = +1
Query: 30 VELMMSGGGGVPVFNLAPNLNVSRLCLGTMTFGEQNTLSQSFQLLDEAYHAGINFFDSAE 89
VELMMSGGGGVPVFNLAPNLNVSRLCLGTMTFGEQNTLSQSFQLLDEAYHAGINFFDSAE
Sbjct: 1 VELMMSGGGGVPVFNLAPNLNVSRLCLGTMTFGEQNTLSQSFQLLDEAYHAGINFFDSAE 180
Query: 90 MYPVPQRAQTWGMSEEYFGHWIKHRNIPRDRLVIATKVAGPSGQMTWIRGGPKSLDATNI 149
MYPVPQRAQTWGMSEEYFGHWIKHRNIPRDRLVIATKVAGPSGQMTWIRGGPKSLDATNI
Sbjct: 181 MYPVPQRAQTWGMSEEYFGHWIKHRNIPRDRLVIATKVAGPSGQMTWIRGGPKSLDATNI 360
Query: 150 SQAIDNSLLRMQLDYIDLYQIHWPDRYVPMFGETEYDPVQQYSSISIDEQLDALSRAVND 209
SQAIDNSLLRMQLDYIDLYQIHWPDRYVPMFGETEYDPVQQYSSISIDEQLDALSRAVND
Sbjct: 361 SQAIDNSLLRMQLDYIDLYQIHWPDRYVPMFGETEYDPVQQYSSISIDEQLDALSRAVND 540
Query: 210 GKIRYIGLSN 219
GKIRYIGLSN
Sbjct: 541 GKIRYIGLSN 570
>TC79432 similar to GP|21689703|gb|AAM67473.1 unknown protein {Arabidopsis
thaliana}, partial (39%)
Length = 751
Score = 150 bits (378), Expect = 9e-37
Identities = 74/157 (47%), Positives = 105/157 (66%), Gaps = 2/157 (1%)
Frame = +2
Query: 48 NLNVSRLCLGTMTFGEQNTLSQSFQLLDEAYHAGINFFDSAEMYPVPQRAQTWGMSEEYF 107
+LN+S + LGTMTFGEQNT +S +L+ A+ GIN D+AE YP+P + +T G ++ Y
Sbjct: 287 DLNISEITLGTMTFGEQNTEKESHDILNYAFENGINALDTAEAYPIPMKKETQGKTDLYI 466
Query: 108 GHWIKHRNIPRDRLVIATKVAGPSGQMTWIRGGPKSL--DATNISQAIDNSLLRMQLDYI 165
W+K ++ RD+++IATKV G S + +++R L DA NI ++++ SL R+ DYI
Sbjct: 467 ASWLKSQS--RDKIIIATKVCGYSERSSYLRDNADILRVDAANIKESVEKSLKRLGTDYI 640
Query: 166 DLYQIHWPDRYVPMFGETEYDPVQQYSSISIDEQLDA 202
DL QIHWPDRYV +FGE YDP + S+ EQL A
Sbjct: 641 DLLQIHWPDRYVALFGEYSYDPSKWRPSVPFVEQLQA 751
>TC86080 similar to PIR|B96632|B96632 hypothetical protein F8A5.20
[imported] - Arabidopsis thaliana, partial (61%)
Length = 1326
Score = 86.7 bits (213), Expect = 1e-17
Identities = 67/239 (28%), Positives = 113/239 (47%), Gaps = 5/239 (2%)
Frame = +3
Query: 51 VSRLCLGTMTF--GEQNTLSQSF--QLLDEAYHAGINFFDSAEMYPVPQRAQTWGMSEEY 106
VS+ LG M G + L + +++ A+ GI FFD+A++Y + G +E
Sbjct: 93 VSKFGLGCMALSGGYNDPLPEEIGISVINHAFSKGITFFDTADVYGLD------GGNEIL 254
Query: 107 FGHWIKHRNIPRDRLVIATKVA-GPSGQMTWIRGGPKSLDATNISQAIDNSLLRMQLDYI 165
G +K +PR+++ +ATK SG I+G P+ + + + SL R+ ++YI
Sbjct: 255 VGKALKQ--LPREKIQVATKFGISRSGGGMGIKGSPEY-----VRSSCEASLKRLNVEYI 413
Query: 166 DLYQIHWPDRYVPMFGETEYDPVQQYSSISIDEQLDALSRAVNDGKIRYIGLSNETPYGL 225
DLY H D VP I++ + L + V +GK++YIGLS +P
Sbjct: 414 DLYYQHRVDTTVP-----------------IEDTVGELKKLVEEGKVKYIGLSEASP--- 533
Query: 226 MKFIQVAEKSSSYPKIVSLQNSYSLLCRTFDSAMAECCHQESISLLAYSPLAMGILSGK 284
++ + I +LQ +SL R ++ + C + I ++ YSPL G +GK
Sbjct: 534 ----DTIRRAHAVHPITALQIEWSLWTRDIENEIVPLCRELGIGIVPYSPLGKGFFAGK 698
>TC86409 similar to PIR|C96632|C96632 hypothetical protein F8A5.21
[imported] - Arabidopsis thaliana, partial (70%)
Length = 1345
Score = 84.3 bits (207), Expect = 6e-17
Identities = 76/256 (29%), Positives = 118/256 (45%), Gaps = 7/256 (2%)
Frame = +2
Query: 49 LNVSRL---CLG-TMTFGEQNTLSQSFQLLDEAYHAGINFFDSAEMYPVPQRAQTWGMSE 104
L VS+L C+G T + + L+ A+ GI FFD+A+ Y A T +E
Sbjct: 152 LEVSKLGYGCMGLTGVYNAAVPEDVAISLIKHAFSKGITFFDTADFYA----AHT---NE 310
Query: 105 EYFGHWIKHRNIPRDRLVIATK---VAGPSGQMTWIRGGPKSLDATNISQAIDNSLLRMQ 161
+ G +K +IPRD++ IATK V SG + + G P+ + + SL R+
Sbjct: 311 VFVGKALK--DIPRDQIQIATKFGIVKMESGNVV-VNGSPEY-----VRSCCEGSLQRLG 466
Query: 162 LDYIDLYQIHWPDRYVPMFGETEYDPVQQYSSISIDEQLDALSRAVNDGKIRYIGLSNET 221
+DYIDLY H D VP I++ + L + V +GKI+YIGLS +
Sbjct: 467 VDYIDLYYQHRIDTTVP-----------------IEDTMGELKKLVEEGKIKYIGLSEAS 595
Query: 222 PYGLMKFIQVAEKSSSYPKIVSLQNSYSLLCRTFDSAMAECCHQESISLLAYSPLAMGIL 281
++ + I ++Q +SL R + + C + I ++ YSPL G
Sbjct: 596 -------TDTIRRAHAVHPITAVQMEWSLWTREIEPDIIPLCRELGIGIVPYSPLGRGFF 754
Query: 282 SGKYFSHGNGPADARL 297
GK + + PAD+ L
Sbjct: 755 GGKAITE-SVPADSFL 799
>TC92605 similar to GP|21689703|gb|AAM67473.1 unknown protein {Arabidopsis
thaliana}, partial (44%)
Length = 850
Score = 58.2 bits (139), Expect(2) = 1e-15
Identities = 34/79 (43%), Positives = 46/79 (58%), Gaps = 4/79 (5%)
Frame = +2
Query: 234 KSSSYPKIVSLQNSYSLLCRT-FDSAMAECCHQES--ISLLAYSPLAMGILSGKYFS-HG 289
K + ++ S+QNSYSLL R+ F+ + E CH ++ I LL+YSPL G LSGKY +
Sbjct: 77 KLRDFQRLFSIQNSYSLLVRSRFEVDLVEVCHPKNCNIGLLSYSPLGGGTLSGKYIDINS 256
Query: 290 NGPADARLNLFKGLLTHCN 308
RLNLF G + N
Sbjct: 257 KAAKSGRLNLFPGYMERYN 313
Score = 42.4 bits (98), Expect(2) = 1e-15
Identities = 19/35 (54%), Positives = 26/35 (74%)
Frame = +1
Query: 208 NDGKIRYIGLSNETPYGLMKFIQVAEKSSSYPKIV 242
N+GK+ Y+G+SNET YG+M+F+ A K PKIV
Sbjct: 1 NEGKVLYLGVSNETSYGVMEFVHAA-KVEGLPKIV 102
>TC77350 similar to PIR|T07394|T07394 probable potassium channel beta chain
KB1 - potato, complete
Length = 1439
Score = 80.1 bits (196), Expect = 1e-15
Identities = 75/255 (29%), Positives = 114/255 (44%), Gaps = 4/255 (1%)
Frame = +1
Query: 49 LNVSRLCLGT-MTFGEQNTLSQSFQLLDEAYHAGINFFDSAEMYPVPQRAQTWGMSEEYF 107
L VS+L G ++FG Q + ++ LL G+NFFD+AE+Y G +EE
Sbjct: 145 LKVSQLSYGAWVSFGNQLDVKEAKSLLQCCRDHGVNFFDNAEVY-------ANGRAEEIM 303
Query: 108 GHWIKHRNIPRDRLVIATKVAGPSGQMTWIRGGP--KSLDATNISQAIDNSLLRMQLDYI 165
G IK + R +V++TK+ W GP K L +I + SL R+ ++Y+
Sbjct: 304 GQAIKSLDWKRSDIVVSTKI-------FWGGQGPNDKGLSRKHIVEGTKASLKRLGMEYV 462
Query: 166 DLYQIHWPDRYVPMFGETEYDPVQQYSSISIDEQLDALSRAVNDGKIRYIGLSNETPYGL 225
D+ H PD P I+E + A++ ++ G Y G S + +
Sbjct: 463 DVVYCHRPDVCTP-----------------IEETVRAMNFVIDQGWAFYWGTSEWSSQQI 591
Query: 226 MKFIQVAEKSSSYPKIVSLQNSYSLLCR-TFDSAMAECCHQESISLLAYSPLAMGILSGK 284
+ VA + +V Q Y+LL R +S I L +SPLA G+L+GK
Sbjct: 592 TEAWAVANRLDLVGPVVE-QPEYNLLNRHKVESEYLPLYSSYGIGLTTWSPLASGVLTGK 768
Query: 285 YFSHGNGPADARLNL 299
Y G P D+R L
Sbjct: 769 Y-KKGVIPPDSRFAL 810
>TC86079 similar to GP|15623896|dbj|BAB67953. putative auxin-induced protein
{Oryza sativa (japonica cultivar-group)}, partial (76%)
Length = 1388
Score = 79.0 bits (193), Expect = 2e-15
Identities = 59/236 (25%), Positives = 107/236 (45%), Gaps = 6/236 (2%)
Frame = +1
Query: 55 CLG-TMTFGEQNTLSQSFQLLDEAYHAGINFFDSAEMYPVPQRAQTWGMSEEYFGHWIKH 113
C+G T + + +++ A+ G+ FFD+A++Y G +E G +K
Sbjct: 121 CMGLTGAYNDPLPEQDGISVINYAFSKGVTFFDTADIYGGS------GANEILLGKALKQ 282
Query: 114 RNIPRDRLVIATKVAGPSGQMT-----WIRGGPKSLDATNISQAIDNSLLRMQLDYIDLY 168
+PR+++ +ATK ++ I+G P+ + + SL R+ ++YIDLY
Sbjct: 283 --LPREKIQLATKFGISRRDVSRLADVTIKGSPEY-----VRSCCEASLKRLDVEYIDLY 441
Query: 169 QIHWPDRYVPMFGETEYDPVQQYSSISIDEQLDALSRAVNDGKIRYIGLSNETPYGLMKF 228
H D +S+SI++ + L + V +GK++YIGLS +P
Sbjct: 442 YQHRID-----------------TSVSIEDTVGELKKLVEEGKVKYIGLSEASP------ 552
Query: 229 IQVAEKSSSYPKIVSLQNSYSLLCRTFDSAMAECCHQESISLLAYSPLAMGILSGK 284
++ + I ++Q +SL R + + C + I ++ YSPL G GK
Sbjct: 553 -DTIRRAHAVHPITAVQIEWSLWTRDIEEEIVPLCRELGIGIVPYSPLGRGFFGGK 717
>TC91056 similar to GP|16555790|emb|CAD10386. L-galactose dehydrogenase
{Arabidopsis thaliana}, complete
Length = 1323
Score = 67.0 bits (162), Expect = 1e-11
Identities = 53/206 (25%), Positives = 94/206 (44%)
Frame = +3
Query: 77 AYHAGINFFDSAEMYPVPQRAQTWGMSEEYFGHWIKHRNIPRDRLVIATKVAGPSGQMTW 136
A+ +GINFFD++ Y +SE+ G +K N+PR ++ATK +
Sbjct: 135 AFQSGINFFDTSPYYGGT-------LSEKVLGKALKALNVPRSEYIVATKCGRYKEGFDF 293
Query: 137 IRGGPKSLDATNISQAIDNSLLRMQLDYIDLYQIHWPDRYVPMFGETEYDPVQQYSSISI 196
A +++++D SL R+QLDY+D+ Q H + E+ + Q +
Sbjct: 294 --------SAERVTRSVDESLERLQLDYVDILQCH----------DIEFGSLDQI----V 407
Query: 197 DEQLDALSRAVNDGKIRYIGLSNETPYGLMKFIQVAEKSSSYPKIVSLQNSYSLLCRTFD 256
+E + AL + GK R+IG+ T L F V ++ V L + + +
Sbjct: 408 NETIPALQKLKEAGKTRFIGI---TGLPLEVFTYVLDRVPPGTLDVILSYCHHSINDSTL 578
Query: 257 SAMAECCHQESISLLAYSPLAMGILS 282
+ + + +++ SPLAMG+L+
Sbjct: 579 EDIVPYLKSKGVGIISASPLAMGLLT 656
>TC85706 similar to GP|21700895|gb|AAM70571.1 At1g60730/F8A5_24 {Arabidopsis
thaliana}, partial (96%)
Length = 1416
Score = 62.4 bits (150), Expect = 2e-10
Identities = 59/238 (24%), Positives = 100/238 (41%), Gaps = 4/238 (1%)
Frame = +2
Query: 49 LNVSRLCLGTMT----FGEQNTLSQSFQLLDEAYHAGINFFDSAEMYPVPQRAQTWGMSE 104
+ VS LG M+ +G L+ A +G+ F D++++Y +E
Sbjct: 137 MEVSLQGLGCMSMSAFYGPPKPEPDMISLIHHAIQSGVTFLDTSDIYGPHT-------NE 295
Query: 105 EYFGHWIKHRNIPRDRLVIATKVAGPSGQMTWIRGGPKSLDATNISQAIDNSLLRMQLDY 164
G +K R+++ +ATK +G + G D + +A + SL R+ +D
Sbjct: 296 VLLGKALKG---VREKVELATKFGVRAGDGKFEICG----DPGYVREACEGSLKRLDIDC 454
Query: 165 IDLYQIHWPDRYVPMFGETEYDPVQQYSSISIDEQLDALSRAVNDGKIRYIGLSNETPYG 224
IDLY H D +P I+ + L + V +GKI+YIGLS +
Sbjct: 455 IDLYYQHRIDTRLP-----------------IEVTIGELKKLVEEGKIKYIGLSEAS--- 574
Query: 225 LMKFIQVAEKSSSYPKIVSLQNSYSLLCRTFDSAMAECCHQESISLLAYSPLAMGILS 282
++ + I ++Q +SL R + + C + I ++AYSPL G S
Sbjct: 575 ----AATIRRAHAVHPITAVQLEWSLWSRDVEEDIIPTCRELGIGIVAYSPLGRGFFS 736
>TC85705 similar to PIR|T12582|T12582 auxin-induced protein - common
sunflower, partial (89%)
Length = 1290
Score = 61.2 bits (147), Expect = 5e-10
Identities = 60/238 (25%), Positives = 102/238 (42%), Gaps = 4/238 (1%)
Frame = +3
Query: 49 LNVSRLCLGTMT----FGEQNTLSQSFQLLDEAYHAGINFFDSAEMYPVPQRAQTWGMSE 104
+ VS LG M+ +G + L+ A +G+ F D++++Y +E
Sbjct: 123 MEVSLQGLGCMSMSAFYGPPKPQTDMIALIHHAIQSGVTFLDTSDIYGPHT-------NE 281
Query: 105 EYFGHWIKHRNIPRDRLVIATKVAGPSGQMTWIRGGPKSLDATNISQAIDNSLLRMQLDY 164
G +K R+++ +ATK + + G D + +A + SL R+ +D
Sbjct: 282 LLLGKALKGG--VREKVELATKFGAKYTEGKFEICG----DPAYVREACEASLKRLDIDC 443
Query: 165 IDLYQIHWPDRYVPMFGETEYDPVQQYSSISIDEQLDALSRAVNDGKIRYIGLSNETPYG 224
IDLY H D +P+ I+I E L + V +GKI+YIGLS +
Sbjct: 444 IDLYYQHRIDTRLPI-------------EITIGE----LKKLVEEGKIKYIGLSEAS--- 563
Query: 225 LMKFIQVAEKSSSYPKIVSLQNSYSLLCRTFDSAMAECCHQESISLLAYSPLAMGILS 282
++ + I ++Q +SL R + + C + I ++AYSPL G S
Sbjct: 564 ----ASTIRRAHAVHPITAVQLEWSLWSRDVEEEIIPTCRELGIGIVAYSPLGRGFFS 725
>TC82833 similar to PIR|T48188|T48188 aldose reductase-like protein -
Arabidopsis thaliana, partial (66%)
Length = 794
Score = 54.7 bits (130), Expect = 5e-08
Identities = 35/133 (26%), Positives = 62/133 (46%)
Frame = +1
Query: 144 LDATNISQAIDNSLLRMQLDYIDLYQIHWPDRYVPMFGETEYDPVQQYSSISIDEQLDAL 203
L + A++N+L +QLDY+DLY +HWP ++ G + + S ++ +
Sbjct: 46 LTPERVRPALNNTLQELQLDYLDLYLVHWP--FLLKDGASRPPKAGEVSEFDMEGVWREM 219
Query: 204 SRAVNDGKIRYIGLSNETPYGLMKFIQVAEKSSSYPKIVSLQNSYSLLCRTFDSAMAECC 263
+ V + +R IG+ N T L K + +A+ P + ++ + M E C
Sbjct: 220 EKLVKENLVRDIGICNFTLTKLDKLVNIAQ---VMPSVCQMEMHPGWR----NDKMLEAC 378
Query: 264 HQESISLLAYSPL 276
+ I + AYSPL
Sbjct: 379 KKNGIHVTAYSPL 417
>TC87522 similar to GP|15215594|gb|AAK91342.1 At1g06690/F4H5_17 {Arabidopsis
thaliana}, partial (46%)
Length = 731
Score = 51.6 bits (122), Expect = 4e-07
Identities = 51/187 (27%), Positives = 81/187 (43%), Gaps = 15/187 (8%)
Frame = +1
Query: 48 NLNVSRLCLGTMTFGEQNTLSQSFQLLDEAYHA-----------GINFFDSAEMYPVPQR 96
+L VS + +G ++G+ T +FQ D A G+ F D+AE+Y
Sbjct: 232 DLKVSTIGIGAWSWGD-TTYWNNFQWNDRNEKAARDAFNTSIDGGLTFIDTAEVYG---S 399
Query: 97 AQTWGM--SEEYFGHWIKHRNI--PRDRLVIATKVAGPSGQMTWIRGGPKSLDATNISQA 152
+G SE G +IK R P + +ATK A P L ++ A
Sbjct: 400 GLAFGAVNSETLLGRFIKERKQKDPNVEVEVATKFAAL----------PWRLGRESVLSA 549
Query: 153 IDNSLLRMQLDYIDLYQIHWPDRYVPMFGETEYDPVQQYSSISIDEQLDALSRAVNDGKI 212
+ +SL R+++ +DLYQ+HWP ++G Y +D L AV G +
Sbjct: 550 LKDSLDRLEMTSVDLYQLHWPG----VWGNEGY--------------IDGLGDAVQKGLV 675
Query: 213 RYIGLSN 219
+ +G+SN
Sbjct: 676 KAVGVSN 696
>TC77142 homologue to GP|9927482|emb|CAC04887.1 unnamed protein product
{Medicago sativa}, complete
Length = 1350
Score = 50.8 bits (120), Expect = 7e-07
Identities = 37/129 (28%), Positives = 56/129 (42%)
Frame = +3
Query: 148 NISQAIDNSLLRMQLDYIDLYQIHWPDRYVPMFGETEYDPVQQYSSISIDEQLDALSRAV 207
++ +A+D +L +QLDY+DLY IHWP V M T + I AL
Sbjct: 285 DVPKALDKTLNDLQLDYLDLYLIHWP---VSMKRGTGEFKAENLDRADIPSTWKALEALY 455
Query: 208 NDGKIRYIGLSNETPYGLMKFIQVAEKSSSYPKIVSLQNSYSLLCRTFDSAMAECCHQES 267
+ GK + IG+SN + L + VA ++ N L + + C +
Sbjct: 456 DSGKAKAIGVSNFSTKKLQDLLDVA-------RVPPAVNQVELHPGWQQAKLHAFCESKG 614
Query: 268 ISLLAYSPL 276
I + YSPL
Sbjct: 615 IHVSGYSPL 641
>TC83181 similar to PIR|T02543|T02543 aldehyde dehydrogenase homolog
At2g37770 - Arabidopsis thaliana, partial (74%)
Length = 655
Score = 49.3 bits (116), Expect = 2e-06
Identities = 34/129 (26%), Positives = 57/129 (43%)
Frame = +3
Query: 148 NISQAIDNSLLRMQLDYIDLYQIHWPDRYVPMFGETEYDPVQQYSSISIDEQLDALSRAV 207
++ A+D +L +QLDY+DLY IHWP PM + + ++ A+
Sbjct: 258 DVPLALDRTLTDLQLDYVDLYLIHWP---APMKKGSVGFKAENLVQPNLASTWKAMEALY 428
Query: 208 NDGKIRYIGLSNETPYGLMKFIQVAEKSSSYPKIVSLQNSYSLLCRTFDSAMAECCHQES 267
+ GK R IG+SN + L ++VA ++ N + + C+ +
Sbjct: 429 DSGKARAIGVSNFSSKKLGDLLEVA-------RVPPAVNQVECHPSWRQDKLRDFCNSKG 587
Query: 268 ISLLAYSPL 276
+ L YSPL
Sbjct: 588 VHLSGYSPL 614
>TC85519 homologue to GP|537298|gb|AAB41556.1|| chalcone reductase {Medicago
sativa subsp. sativa}, complete
Length = 1167
Score = 48.9 bits (115), Expect = 3e-06
Identities = 54/214 (25%), Positives = 84/214 (39%), Gaps = 6/214 (2%)
Frame = +1
Query: 76 EAYHAGINFFDSAEMYPVPQRAQTWGMSEEYFGHWIKHRN----IPRDRLVIATKVAGPS 131
EA G FD+A Y SE+ G +K + R L + +K+
Sbjct: 202 EAIKQGYRHFDTAAAYG----------SEQALGEALKEAIELGLVTRQDLFVTSKL---- 339
Query: 132 GQMTWIRGGPKSLDATNISQAIDNSLLRMQLDYIDLYQIHWPDRYVPMFGETEYDPVQQY 191
W+ L + A+ SL +QLDY+DLY IHWP P G+ + P+
Sbjct: 340 ----WVTENHPHL----VIPALQKSLKTLQLDYLDLYLIHWPLSSQP--GKFTF-PIDVA 486
Query: 192 SSISIDEQ--LDALSRAVNDGKIRYIGLSNETPYGLMKFIQVAEKSSSYPKIVSLQNSYS 249
+ D + +++ + G + IG+SN + L + VA I+ N
Sbjct: 487 DLLPFDVKGVWESMEEGLKLGLTKAIGVSNFSVKKLENLLSVA-------TILPAVNQVE 645
Query: 250 LLCRTFDSAMAECCHQESISLLAYSPLAMGILSG 283
+ + E C+ I L A+SPL G G
Sbjct: 646 MNLAWQQKKLREFCNANGIVLTAFSPLRKGASRG 747
>CA857968 similar to PIR|T48188|T48 aldose reductase-like protein -
Arabidopsis thaliana, partial (66%)
Length = 775
Score = 47.8 bits (112), Expect = 6e-06
Identities = 31/117 (26%), Positives = 54/117 (45%)
Frame = +2
Query: 160 MQLDYIDLYQIHWPDRYVPMFGETEYDPVQQYSSISIDEQLDALSRAVNDGKIRYIGLSN 219
+QLDY+DLY +HWP ++ G + + S ++ + + V + +R IG+ N
Sbjct: 8 LQLDYLDLYLVHWP--FLLKDGASRPPKAGEVSEFDMEGVWREMEKLVKENLVRDIGICN 181
Query: 220 ETPYGLMKFIQVAEKSSSYPKIVSLQNSYSLLCRTFDSAMAECCHQESISLLAYSPL 276
T L K + +A+ P + ++ + M E C + I + AYSPL
Sbjct: 182 FTLTKLDKLVNIAQ---VMPSVCQMEMHPGWR----NDKMLEACKKNGIHVTAYSPL 331
>TC78085 similar to GP|9927482|emb|CAC04887.1 unnamed protein product
{Medicago sativa}, partial (80%)
Length = 1030
Score = 47.8 bits (112), Expect = 6e-06
Identities = 38/135 (28%), Positives = 57/135 (42%)
Frame = +2
Query: 148 NISQAIDNSLLRMQLDYIDLYQIHWPDRYVPMFGETEYDPVQQYSSISIDEQLDALSRAV 207
++ +A D +L +QLDY+DLY IHWP V + P I A+
Sbjct: 338 DVPKAFDRTLRDLQLDYLDLYLIHWP---VSIKNGHLTKP-------DIPSTWKAMEALY 487
Query: 208 NDGKIRYIGLSNETPYGLMKFIQVAEKSSSYPKIVSLQNSYSLLCRTFDSAMAECCHQES 267
+ GK R IG+SN + L + V ++ N L + + C +
Sbjct: 488 DSGKARAIGVSNFSVKKLQDLLDVG-------RVPPAVNQVELHPQLQQPNLHTFCKSKG 646
Query: 268 ISLLAYSPLAMGILS 282
+ L AYSPL G+ S
Sbjct: 647 VHLSAYSPLGKGLES 691
>TC78138 similar to GP|2792155|emb|CAA11226.1 chalcone reductase {Sesbania
rostrata}, partial (83%)
Length = 1289
Score = 46.6 bits (109), Expect = 1e-05
Identities = 53/216 (24%), Positives = 90/216 (41%)
Frame = +2
Query: 61 FGEQNTLSQSFQLLDEAYHAGINFFDSAEMYPVPQRAQTWGMSEEYFGHWIKHRNIPRDR 120
FG T Q Q++ +A G FD+A +Y + +S+ +K+R D
Sbjct: 116 FGTGTTPPQ--QIMLDAIDIGYRHFDTAALYGTEEPLGQ-AVSKALELGLVKNR----DE 274
Query: 121 LVIATKVAGPSGQMTWIRGGPKSLDATNISQAIDNSLLRMQLDYIDLYQIHWPDRYVPMF 180
L I +K+ W L + A+ +L + L+Y+DLY IHWP R +
Sbjct: 275 LFITSKL--------WCTDAQHDL----VLPALKTTLKNLGLEYVDLYLIHWPVR-LKQD 415
Query: 181 GETEYDPVQQYSSISIDEQLDALSRAVNDGKIRYIGLSNETPYGLMKFIQVAEKSSSYPK 240
E+ + I +A+ G + IG+SN +G+ K + E + P
Sbjct: 416 AESLKFKKEDMIPFDIKGTWEAMEECYRLGLAKSIGVSN---FGVKKLSILLENAEIAPA 586
Query: 241 IVSLQNSYSLLCRTFDSAMAECCHQESISLLAYSPL 276
+ ++ + S + E C Q+ I + A+SPL
Sbjct: 587 VNQVEMNPSWQ----QGKLREFCKQKGIHVSAWSPL 682
>TC87245 similar to GP|10334991|gb|AAG15839.2 NADPH-dependent mannose
6-phosphate reductase {Orobanche ramosa}, partial (98%)
Length = 1196
Score = 45.8 bits (107), Expect = 2e-05
Identities = 37/153 (24%), Positives = 68/153 (44%), Gaps = 11/153 (7%)
Frame = +3
Query: 145 DATNISQAIDNSLLRMQLDYIDLYQIHWP-----------DRYVPMFGETEYDPVQQYSS 193
D ++ +A +SL ++QLDY+DLY +H+P D + G + D ++
Sbjct: 267 DHGHVVEACKDSLKKLQLDYLDLYLVHFPVATRHTGVGTTDSALGEDGVLDID-----TT 431
Query: 194 ISIDEQLDALSRAVNDGKIRYIGLSNETPYGLMKFIQVAEKSSSYPKIVSLQNSYSLLCR 253
IS++ A+ V+ G +R IG+SN I + +Y KI N
Sbjct: 432 ISLETTWHAMEGLVSSGLVRSIGISNYD-------IFLTRDCLAYSKIKPAVNQIETHPY 590
Query: 254 TFDSAMAECCHQESISLLAYSPLAMGILSGKYF 286
++ + C + I + A++PL + ++F
Sbjct: 591 FQRESLVKFCQKHGICVTAHTPLGGAAANKEWF 689
>TC85521 homologue to GP|537298|gb|AAB41556.1|| chalcone reductase {Medicago
sativa subsp. sativa}, complete
Length = 1508
Score = 45.1 bits (105), Expect = 4e-05
Identities = 41/148 (27%), Positives = 63/148 (41%), Gaps = 6/148 (4%)
Frame = +3
Query: 142 KSLDATNISQAIDNSLL----RMQLDYIDLYQIHWPDRYVPMFGETEYDPVQQYSSISID 197
+ D+T I D LL +QLDY+DLY IHWP P G+ + P+ + D
Sbjct: 573 RMFDSTMIKINFDLDLLISCRTLQLDYLDLYLIHWPLSSQP--GKFTF-PIDVADLLPFD 743
Query: 198 EQ--LDALSRAVNDGKIRYIGLSNETPYGLMKFIQVAEKSSSYPKIVSLQNSYSLLCRTF 255
+ +++ + G + IG+SN + L + VA I+ N +
Sbjct: 744 VKGVWESMEEGLKLGLTKAIGVSNFSVKKLENLLSVA-------TILPAVNQVEMNLAWQ 902
Query: 256 DSAMAECCHQESISLLAYSPLAMGILSG 283
+ E C+ I L A+SPL G G
Sbjct: 903 QKKLREFCNANGIVLTAFSPLRKGASRG 986
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.323 0.138 0.422
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 11,579,950
Number of Sequences: 36976
Number of extensions: 164375
Number of successful extensions: 912
Number of sequences better than 10.0: 54
Number of HSP's better than 10.0 without gapping: 892
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 897
length of query: 337
length of database: 9,014,727
effective HSP length: 97
effective length of query: 240
effective length of database: 5,428,055
effective search space: 1302733200
effective search space used: 1302733200
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.5 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (22.0 bits)
S2: 59 (27.3 bits)
Medicago: description of AC141106.8


