

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC140104.6 - phase: 0
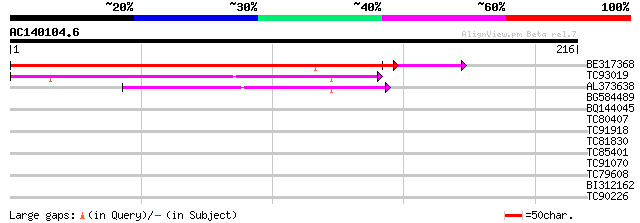
(216 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
BE317368 189 3e-49
TC93019 weakly similar to PIR|E84642|E84642 hypothetical protein... 52 2e-07
AL373638 similar to PIR|S27776|S277 80K protein (allele C1B) - B... 44 5e-05
BG584489 similar to PIR|T47792|T4 hypothetical protein F17J16.15... 33 0.064
BQ144045 similar to PIR|T51947|T51 probable transcription factor... 32 0.24
TC80407 similar to GP|10177633|dbj|BAB10781. gene_id:K18B18.4~un... 31 0.41
TC91918 homologue to GP|9369375|gb|AAF87124.1| F10A5.29 {Arabido... 28 2.1
TC81830 similar to GP|15982206|emb|CAC83608. Na+/H+ antiporter ... 27 4.6
TC85401 homologue to GP|13160142|emb|CAC32462. sucrose synthase ... 27 4.6
TC91070 weakly similar to GP|1086592|gb|AAA82270.1| C. elegans R... 27 4.6
TC79608 similar to GP|20147231|gb|AAM10330.1 At1g68570/F24J5_7 {... 27 6.0
BI312162 similar to GP|4150963|emb DsPTP1 protein {Arabidopsis t... 27 7.8
TC90226 similar to GP|9758397|dbj|BAB08884.1 signal recognition ... 27 7.8
>BE317368
Length = 621
Score = 189 bits (481), Expect(2) = 3e-49
Identities = 96/149 (64%), Positives = 114/149 (76%), Gaps = 1/149 (0%)
Frame = +2
Query: 1 MASKESLPNSSENLGKAKWDTFSTKMLLDICMVEIHKCGKLGIAFRNKKWEEIRDEYNKR 60
MASKESL + E KAKWD STKMLL+IC EI K K GIAF+NKKWEEIR E+N R
Sbjct: 65 MASKESLICNHEIPEKAKWDPISTKMLLNICGDEICKTRKPGIAFKNKKWEEIRVEFNNR 244
Query: 61 ANKNYTQKQLKNRMDTLRGEWTIWKQLLGKETGLGWNHHIGNIDADSAWWDAKIR-NVKY 119
A +N+TQKQLKNRMD+LR +W +WKQL+GKET + N+H I+A+SAWWDAKIR N K+
Sbjct: 245 AKRNHTQKQLKNRMDSLRADWILWKQLMGKETHVELNNHTKTIEANSAWWDAKIRENAKF 424
Query: 120 AKFRFQGLEFCDELEFIFGEIVATSQCAC 148
A+FR+QGLEF DELE IFGE A + C
Sbjct: 425 ARFRYQGLEFRDELEHIFGEPKANHKSKC 511
Score = 22.3 bits (46), Expect(2) = 3e-49
Identities = 13/32 (40%), Positives = 19/32 (58%)
Frame = +3
Query: 143 TSQCACAPAMGVPLESTGKNTTADVAREIIES 174
TSQ A P +G+ +ES + +V +E IES
Sbjct: 498 TSQNAQTPIVGLSIESR*QKY*FNVPQEAIES 593
>TC93019 weakly similar to PIR|E84642|E84642 hypothetical protein At2g24960
[imported] - Arabidopsis thaliana, partial (8%)
Length = 718
Score = 51.6 bits (122), Expect = 2e-07
Identities = 36/145 (24%), Positives = 64/145 (43%), Gaps = 3/145 (2%)
Frame = +2
Query: 1 MASKESLPNSSENL--GKAKWDTFSTKMLLDICMVEIHKCGKLGIAFRNKKWEEIRDEYN 58
M E +SS NL +A W + L++ + +HK K G F W ++ +++N
Sbjct: 134 MQGNEIGDDSSINLDNSRANWTPSQDQYFLELLLSHVHKGNKTGKVFTRLAWADMTEQFN 313
Query: 59 KRANKNYTQKQLKNRMDTLRGEWTIWKQLLGKETGLGWNHHIGNIDADSAWWDAKIRNVK 118
+ Y LKNR + ++ K + + G W+ + I A+ W+ I+
Sbjct: 314 SKFGFKYDVDVLKNRYKRFKKQYYEIK-AMASQNGFQWDGRLNTITANDKTWEEYIKAHP 490
Query: 119 YAK-FRFQGLEFCDELEFIFGEIVA 142
A+ FR + ++L I+G VA
Sbjct: 491 DAQVFRKKLFPCYNDLCIIYGHTVA 565
>AL373638 similar to PIR|S27776|S277 80K protein (allele C1B) - Babesia
bovis, partial (3%)
Length = 442
Score = 43.9 bits (102), Expect = 5e-05
Identities = 30/103 (29%), Positives = 47/103 (45%), Gaps = 1/103 (0%)
Frame = +2
Query: 44 AFRNKKWEEIRDEYNKRANKNYTQKQLKNRMDTLRGEWTIWKQLLGKETGLGWNHHIGNI 103
+F K W+ I D ++ + + +++LKNR LR ++ I K +L E W+ G I
Sbjct: 20 SFNKKAWKHICDGFHNKTGLKWDKEKLKNRHSVLRRQYAIVKPIL-DEGDFVWDEATGAI 196
Query: 104 DADSAWWDAKIRNVKYAK-FRFQGLEFCDELEFIFGEIVATSQ 145
A+ W I+N A+ + G EL IF E Q
Sbjct: 197 IANDEIWAEYIKNNPDAETVKSGGCSIFKELCTIFSEAATNGQ 325
>BG584489 similar to PIR|T47792|T4 hypothetical protein F17J16.150 -
Arabidopsis thaliana, partial (12%)
Length = 823
Score = 33.5 bits (75), Expect = 0.064
Identities = 15/49 (30%), Positives = 28/49 (56%)
Frame = +3
Query: 80 EWTIWKQLLGKETGLGWNHHIGNIDADSAWWDAKIRNVKYAKFRFQGLE 128
+W+ WK+ +G G+G I + + +WWD + ++KY+ R + LE
Sbjct: 234 DWSDWKRWMGNRGGIG----IPSDKSWESWWDEENEHLKYSNVRGKILE 368
>BQ144045 similar to PIR|T51947|T51 probable transcription factor HUA2
[imported] - Arabidopsis thaliana, partial (1%)
Length = 1172
Score = 31.6 bits (70), Expect = 0.24
Identities = 14/34 (41%), Positives = 19/34 (55%)
Frame = +2
Query: 62 NKNYTQKQLKNRMDTLRGEWTIWKQLLGKETGLG 95
N+ T KQ KN +D + G W +W +LG G G
Sbjct: 317 NRTQTIKQNKNSLDEVEGVWRVW*GMLGCFGGSG 418
>TC80407 similar to GP|10177633|dbj|BAB10781. gene_id:K18B18.4~unknown
protein {Arabidopsis thaliana}, partial (7%)
Length = 1159
Score = 30.8 bits (68), Expect = 0.41
Identities = 17/49 (34%), Positives = 28/49 (56%)
Frame = -1
Query: 148 CAPAMGVPLESTGKNTTADVAREIIESDDELNIDELSPMENTQSKNKRK 196
C PA VPLE+ + D++ ++ + E + + LSPM +T +N RK
Sbjct: 544 CFPAQMVPLETKSLQNSPDLSIKVTKEMFETS-NSLSPMASTIRRNTRK 401
>TC91918 homologue to GP|9369375|gb|AAF87124.1| F10A5.29 {Arabidopsis
thaliana}, partial (38%)
Length = 1171
Score = 28.5 bits (62), Expect = 2.1
Identities = 17/40 (42%), Positives = 27/40 (67%), Gaps = 1/40 (2%)
Frame = +2
Query: 168 AREIIESDDELNIDELS-PMENTQSKNKRKVSPRMRKQQR 206
AR++ SD +LN+ ELS P +NT S + P+M++QQ+
Sbjct: 254 ARKLYGSDAKLNLPELSTPPQNTTS-SPSPTPPQMQQQQQ 370
>TC81830 similar to GP|15982206|emb|CAC83608. Na+/H+ antiporter isoform 2
{Lycopersicon esculentum}, partial (45%)
Length = 793
Score = 27.3 bits (59), Expect = 4.6
Identities = 12/37 (32%), Positives = 17/37 (45%)
Frame = +1
Query: 73 RMDTLRGEWTIWKQLLGKETGLGWNHHIGNIDADSAW 109
RM +L W+IW LL + H G +D + W
Sbjct: 325 RMSSLVLTWSIWSDLLIERYLQNTKRHFGIVDFEGQW 435
>TC85401 homologue to GP|13160142|emb|CAC32462. sucrose synthase isoform 3
{Pisum sativum}, complete
Length = 2846
Score = 27.3 bits (59), Expect = 4.6
Identities = 12/38 (31%), Positives = 21/38 (54%)
Frame = +2
Query: 50 WEEIRDEYNKRANKNYTQKQLKNRMDTLRGEWTIWKQL 87
W++I KR ++ YT + +R+ TL G + WK +
Sbjct: 2342 WDKISHGGLKRIHEKYTWQIYSDRLLTLTGVYGFWKHV 2455
>TC91070 weakly similar to GP|1086592|gb|AAA82270.1| C. elegans RSP-6
protein (corresponding sequence C33H5.12a)
{Caenorhabditis elegans}, partial (17%)
Length = 859
Score = 27.3 bits (59), Expect = 4.6
Identities = 16/46 (34%), Positives = 25/46 (53%), Gaps = 2/46 (4%)
Frame = +2
Query: 60 RANKNYTQKQLKNRMDTLRGEWTIWKQLLGKETGLGWNHH--IGNI 103
R +K Q ++K + T R T KQL+ + + WNHH +GN+
Sbjct: 665 RVSKYSKQLKMKEKTRTWRFTLTFLKQLMARV--MMWNHHFLLGNL 796
>TC79608 similar to GP|20147231|gb|AAM10330.1 At1g68570/F24J5_7 {Arabidopsis
thaliana}, partial (59%)
Length = 1489
Score = 26.9 bits (58), Expect = 6.0
Identities = 9/17 (52%), Positives = 12/17 (69%), Gaps = 2/17 (11%)
Frame = +1
Query: 96 WNHHIGNIDADSA--WW 110
WN HIGN+D ++ WW
Sbjct: 4 WNPHIGNVDLNNCIHWW 54
>BI312162 similar to GP|4150963|emb DsPTP1 protein {Arabidopsis thaliana},
partial (72%)
Length = 761
Score = 26.6 bits (57), Expect = 7.8
Identities = 9/22 (40%), Positives = 14/22 (62%)
Frame = -1
Query: 30 ICMVEIHKCGKLGIAFRNKKWE 51
+C+V IH C KLG+ K++
Sbjct: 689 VCIVAIHSCNKLGLPHLRMKFQ 624
>TC90226 similar to GP|9758397|dbj|BAB08884.1 signal recognition particle
68kD protein-like {Arabidopsis thaliana}, partial (40%)
Length = 845
Score = 26.6 bits (57), Expect = 7.8
Identities = 11/27 (40%), Positives = 17/27 (62%)
Frame = -3
Query: 59 KRANKNYTQKQLKNRMDTLRGEWTIWK 85
K+++ Y Q N++D+LR W IWK
Sbjct: 495 KQSSPLYCLAQPTNQIDSLRCIWAIWK 415
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.317 0.132 0.402
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 6,470,200
Number of Sequences: 36976
Number of extensions: 84561
Number of successful extensions: 432
Number of sequences better than 10.0: 26
Number of HSP's better than 10.0 without gapping: 429
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 430
length of query: 216
length of database: 9,014,727
effective HSP length: 92
effective length of query: 124
effective length of database: 5,612,935
effective search space: 696003940
effective search space used: 696003940
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 56 (26.2 bits)
Medicago: description of AC140104.6


