

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC140104.12 - phase: 0
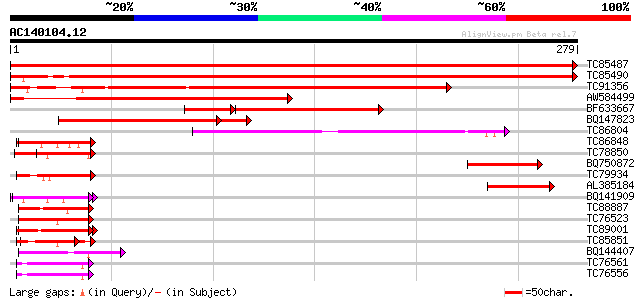
(279 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC85487 homologue to SP|P49688|RS2_ARATH 40S ribosomal protein S... 565 e-162
TC85490 similar to GP|18146720|dbj|BAB82426. ribosomal protein S... 526 e-150
TC91356 similar to GP|20151585|gb|AAM11152.1 LD24077p {Drosophil... 285 2e-77
AW584499 similar to GP|21536514|gb 40S ribosomal protein S2 homo... 215 2e-56
BF633667 similar to PIR|A31139|A311 ribosomal protein S2 - mouse... 136 2e-40
BQ147823 similar to GP|18146720|dbj ribosomal protein S2 {Arabid... 149 5e-39
TC86804 similar to GP|5725342|emb|CAA63650.1 ribosomal protein S... 79 2e-15
TC86848 similar to GP|10177413|dbj|BAB10544. fibrillarin-like {A... 52 3e-07
TC78850 homologue to GP|10177413|dbj|BAB10544. fibrillarin-like ... 48 4e-06
BQ750872 similar to PIR|A36363|R3BY ribosomal protein S2.e cyto... 47 8e-06
TC79934 similar to GP|20259633|gb|AAM14173.1 putative GAR1 prote... 45 2e-05
AL385184 weakly similar to SP|Q90YS3|RS2_I 40S ribosomal protein... 45 4e-05
BQ141909 weakly similar to GP|14253159|emb VMP3 protein {Volvox ... 44 5e-05
TC88887 weakly similar to GP|6523547|emb|CAB62280.1 hydroxyproli... 44 7e-05
TC76523 homologue to PIR|T09648|T09648 nucleolin homolog nuM1 - ... 44 9e-05
TC89001 similar to GP|5726567|gb|AAD48471.1| glycine-rich RNA-bi... 44 9e-05
TC85851 similar to PIR|T12180|T12180 probable transcription fact... 44 9e-05
BQ144407 weakly similar to GP|6523547|emb| hydroxyproline-rich g... 43 1e-04
TC76561 similar to PIR|S71779|S71779 glycine-rich RNA-binding pr... 41 6e-04
TC76556 similar to PIR|S71779|S71779 glycine-rich RNA-binding pr... 41 6e-04
>TC85487 homologue to SP|P49688|RS2_ARATH 40S ribosomal protein S2.
[Mouse-ear cress] {Arabidopsis thaliana}, partial (88%)
Length = 1114
Score = 565 bits (1455), Expect = e-162
Identities = 279/279 (100%), Positives = 279/279 (100%)
Frame = +1
Query: 1 MAERGGDRGGFGSGFGGRGGDRGGRGGRGDRGRGGRRRAGGRREEEEKWVPVTKLGRLVK 60
MAERGGDRGGFGSGFGGRGGDRGGRGGRGDRGRGGRRRAGGRREEEEKWVPVTKLGRLVK
Sbjct: 73 MAERGGDRGGFGSGFGGRGGDRGGRGGRGDRGRGGRRRAGGRREEEEKWVPVTKLGRLVK 252
Query: 61 EGKIRSLEQIYLHSLPIKEHQIIDTLVGPTLKDEVMKIMPVQKQTRAGQRTRFKAFVVVG 120
EGKIRSLEQIYLHSLPIKEHQIIDTLVGPTLKDEVMKIMPVQKQTRAGQRTRFKAFVVVG
Sbjct: 253 EGKIRSLEQIYLHSLPIKEHQIIDTLVGPTLKDEVMKIMPVQKQTRAGQRTRFKAFVVVG 432
Query: 121 DNNGHVGLGVKCSKEVATAIRGAIILAKLSVIPVRRGYWGNKIGKPHTVPCKVTGKCGSV 180
DNNGHVGLGVKCSKEVATAIRGAIILAKLSVIPVRRGYWGNKIGKPHTVPCKVTGKCGSV
Sbjct: 433 DNNGHVGLGVKCSKEVATAIRGAIILAKLSVIPVRRGYWGNKIGKPHTVPCKVTGKCGSV 612
Query: 181 TVRMVPAPRGSGIVAARVPKKVLQFAGIDDVFTSSRGSTKTLGNFVKATFDCLMKTYGFL 240
TVRMVPAPRGSGIVAARVPKKVLQFAGIDDVFTSSRGSTKTLGNFVKATFDCLMKTYGFL
Sbjct: 613 TVRMVPAPRGSGIVAARVPKKVLQFAGIDDVFTSSRGSTKTLGNFVKATFDCLMKTYGFL 792
Query: 241 TPEFWKETRFSKSPFQEYTDLLAKPTAKALILEEERVEA 279
TPEFWKETRFSKSPFQEYTDLLAKPTAKALILEEERVEA
Sbjct: 793 TPEFWKETRFSKSPFQEYTDLLAKPTAKALILEEERVEA 909
>TC85490 similar to GP|18146720|dbj|BAB82426. ribosomal protein S2
{Arabidopsis thaliana}, partial (89%)
Length = 1252
Score = 526 bits (1356), Expect = e-150
Identities = 267/281 (95%), Positives = 269/281 (95%), Gaps = 2/281 (0%)
Frame = +1
Query: 1 MAERG--GDRGGFGSGFGGRGGDRGGRGGRGDRGRGGRRRAGGRREEEEKWVPVTKLGRL 58
MA+RG GDR GFG GFG RG RGGRGG DRGRGGRRR GRREEEEKWVPVTKLGRL
Sbjct: 58 MADRGATGDRAGFGRGFGDRG--RGGRGG--DRGRGGRRRGAGRREEEEKWVPVTKLGRL 225
Query: 59 VKEGKIRSLEQIYLHSLPIKEHQIIDTLVGPTLKDEVMKIMPVQKQTRAGQRTRFKAFVV 118
VKEGKIRSLEQIYLHSLPIKEHQIIDTLVGPTLKDEVMKIMPVQKQTRAGQRTRFKAFVV
Sbjct: 226 VKEGKIRSLEQIYLHSLPIKEHQIIDTLVGPTLKDEVMKIMPVQKQTRAGQRTRFKAFVV 405
Query: 119 VGDNNGHVGLGVKCSKEVATAIRGAIILAKLSVIPVRRGYWGNKIGKPHTVPCKVTGKCG 178
VGDNNGHVGLGVKCSKEVATAIRGAIILAKLSVIPVRRGYWGNKIGKPHTVPCKVTGKCG
Sbjct: 406 VGDNNGHVGLGVKCSKEVATAIRGAIILAKLSVIPVRRGYWGNKIGKPHTVPCKVTGKCG 585
Query: 179 SVTVRMVPAPRGSGIVAARVPKKVLQFAGIDDVFTSSRGSTKTLGNFVKATFDCLMKTYG 238
SVTVRMVPAPRGSGIVAARVPKKVLQFAGIDDVFTSSRGSTKTLGNFVKATFDCLMKTYG
Sbjct: 586 SVTVRMVPAPRGSGIVAARVPKKVLQFAGIDDVFTSSRGSTKTLGNFVKATFDCLMKTYG 765
Query: 239 FLTPEFWKETRFSKSPFQEYTDLLAKPTAKALILEEERVEA 279
FLTPEFWKETRFSKSPFQEYTD+LAKPT KALILEEERVEA
Sbjct: 766 FLTPEFWKETRFSKSPFQEYTDMLAKPTTKALILEEERVEA 888
>TC91356 similar to GP|20151585|gb|AAM11152.1 LD24077p {Drosophila
melanogaster}, partial (71%)
Length = 698
Score = 285 bits (728), Expect = 2e-77
Identities = 149/220 (67%), Positives = 177/220 (79%), Gaps = 3/220 (1%)
Frame = +2
Query: 1 MAERGGD--RGGFGSGFGGRGGDRGGRGGRGDRGRG-GRRRAGGRREEEEKWVPVTKLGR 57
MA+RG RGG GFG RGGDRG RGRG GR R GG+ EE+E W PVTKLGR
Sbjct: 74 MADRGASAPRGG---GFGSRGGDRG-------RGRGRGRGRRGGKSEEKE-WQPVTKLGR 220
Query: 58 LVKEGKIRSLEQIYLHSLPIKEHQIIDTLVGPTLKDEVMKIMPVQKQTRAGQRTRFKAFV 117
LVK GKI+S+E+IYLHSLPIKE+QI+D + P LKDEVMKI PVQKQTRAGQRTRFKA V
Sbjct: 221 LVKAGKIQSMEEIYLHSLPIKEYQIVDYFL-PKLKDEVMKIKPVQKQTRAGQRTRFKAIV 397
Query: 118 VVGDNNGHVGLGVKCSKEVATAIRGAIILAKLSVIPVRRGYWGNKIGKPHTVPCKVTGKC 177
++GD+ GHVGLG+K SKEVATAIR AII+AKLSVIPVR GYWG +G PH++P K +GKC
Sbjct: 398 LIGDSEGHVGLGIKTSKEVATAIRAAIIIAKLSVIPVRLGYWGTNLGVPHSLPTKESGKC 577
Query: 178 GSVTVRMVPAPRGSGIVAARVPKKVLQFAGIDDVFTSSRG 217
GSVTVR++PAPRG+G+VA+ K++LQ AG++D +TSS G
Sbjct: 578 GSVTVRLIPAPRGTGLVASPAVKRLLQLAGVEDAYTSSAG 697
>AW584499 similar to GP|21536514|gb 40S ribosomal protein S2 homolog
{Arabidopsis thaliana}, partial (40%)
Length = 664
Score = 215 bits (547), Expect = 2e-56
Identities = 113/139 (81%), Positives = 114/139 (81%)
Frame = +2
Query: 1 MAERGGDRGGFGSGFGGRGGDRGGRGGRGDRGRGGRRRAGGRREEEEKWVPVTKLGRLVK 60
MAERGGD RGGRRRAGGRREEEEKWVPVTKLGRLVK
Sbjct: 323 MAERGGD-------------------------RGGRRRAGGRREEEEKWVPVTKLGRLVK 427
Query: 61 EGKIRSLEQIYLHSLPIKEHQIIDTLVGPTLKDEVMKIMPVQKQTRAGQRTRFKAFVVVG 120
EGKIRSLEQIYLHSLPIKEHQIIDTLVGPTLKDEVMKIMPVQKQTRAGQRTRFKAFVVVG
Sbjct: 428 EGKIRSLEQIYLHSLPIKEHQIIDTLVGPTLKDEVMKIMPVQKQTRAGQRTRFKAFVVVG 607
Query: 121 DNNGHVGLGVKCSKEVATA 139
DNNGHVGLGVKC+KEVATA
Sbjct: 608 DNNGHVGLGVKCNKEVATA 664
>BF633667 similar to PIR|A31139|A311 ribosomal protein S2 - mouse (fragment),
partial (44%)
Length = 298
Score = 136 bits (342), Expect(2) = 2e-40
Identities = 64/73 (87%), Positives = 69/73 (93%)
Frame = +3
Query: 112 RFKAFVVVGDNNGHVGLGVKCSKEVATAIRGAIILAKLSVIPVRRGYWGNKIGKPHTVPC 171
RFKAFVVVGD+NGHVGLGVKC+KEVATAIRGAIILAK+SV+PVRRGYWGNKIG HTVP
Sbjct: 78 RFKAFVVVGDHNGHVGLGVKCAKEVATAIRGAIILAKMSVVPVRRGYWGNKIGSVHTVPA 257
Query: 172 KVTGKCGSVTVRM 184
KVTGKCGSV VR+
Sbjct: 258 KVTGKCGSVIVRL 296
Score = 47.4 bits (111), Expect(2) = 2e-40
Identities = 22/25 (88%), Positives = 24/25 (96%)
Frame = +2
Query: 87 VGPTLKDEVMKIMPVQKQTRAGQRT 111
+G +LKDEVMKIMPVQKQTRAGQRT
Sbjct: 2 LGASLKDEVMKIMPVQKQTRAGQRT 76
>BQ147823 similar to GP|18146720|dbj ribosomal protein S2 {Arabidopsis
thaliana}, partial (31%)
Length = 695
Score = 149 bits (377), Expect(2) = 5e-39
Identities = 73/80 (91%), Positives = 75/80 (93%)
Frame = +3
Query: 25 RGGRGDRGRGGRRRAGGRREEEEKWVPVTKLGRLVKEGKIRSLEQIYLHSLPIKEHQIID 84
+ GRGDRGRGGRR AGGRREEEEKWVPVTKLGRLVKE KIRSLEQIYLHSLPIKEHQIID
Sbjct: 411 KSGRGDRGRGGRRXAGGRREEEEKWVPVTKLGRLVKEXKIRSLEQIYLHSLPIKEHQIID 590
Query: 85 TLVGPTLKDEVMKIMPVQKQ 104
TLVGPTLKD VMKIMPVQ +
Sbjct: 591 TLVGPTLKDXVMKIMPVQNK 650
Score = 28.9 bits (63), Expect(2) = 5e-39
Identities = 14/17 (82%), Positives = 14/17 (82%)
Frame = +2
Query: 103 KQTRAGQRTRFKAFVVV 119
KQTRAGQ T KAFVVV
Sbjct: 644 KQTRAGQXTPXKAFVVV 694
>TC86804 similar to GP|5725342|emb|CAA63650.1 ribosomal protein S5 {Spinacia
oleracea}, partial (75%)
Length = 1353
Score = 79.0 bits (193), Expect = 2e-15
Identities = 54/165 (32%), Positives = 86/165 (51%), Gaps = 9/165 (5%)
Frame = +1
Query: 91 LKDEVMKIMPVQKQTRAGQRTRFKAFVVVGDNNGHVGLGVKCSKEVATAIRGAIILAKLS 150
L++ V+++ V K + G++ RF+A VVVGD G VG+GV +KEV +A++ + I A+ +
Sbjct: 559 LEERVIQVRRVTKVVKGGKQMRFRAIVVVGDKQGRVGVGVGKAKEVVSAVQKSAINARRN 738
Query: 151 VIPVRRGYWGNKIGKPHTVPCKVTGKCGSVTVRMVPAPRGSGIVAARVPKKVLQFAGIDD 210
+I V + K T P + G G+ V + PA G+G++A + VL+ AG+++
Sbjct: 739 LIRV-------PMTKYLTFPHRADGDYGASKVMLRPASPGTGVIAGGSVRIVLEMAGVEN 897
Query: 211 VFTSSRGSTKTLGNFVKATFDCL--MKTY-------GFLTPEFWK 246
GS L N +AT + MK Y G E WK
Sbjct: 898 ALGKQLGSDNALNN-ARATIAAVQKMKQYSEVAEERGIPMEELWK 1029
>TC86848 similar to GP|10177413|dbj|BAB10544. fibrillarin-like {Arabidopsis
thaliana}, partial (92%)
Length = 1250
Score = 51.6 bits (122), Expect = 3e-07
Identities = 31/50 (62%), Positives = 31/50 (62%), Gaps = 11/50 (22%)
Frame = +3
Query: 4 RGGDRGGFGSG---FGGRGGDR--------GGRGGRGDRGRGGRRRAGGR 42
RGGDRGG G G FGGRGGDR GGRGG G GRGG R GGR
Sbjct: 105 RGGDRGGRGGGRGGFGGRGGDRGTPFKARGGGRGGGGRGGRGG-GRGGGR 251
Score = 48.5 bits (114), Expect = 3e-06
Identities = 28/44 (63%), Positives = 28/44 (63%), Gaps = 6/44 (13%)
Frame = +3
Query: 5 GGDRGGFGSGFGGRGGDRGGRGGR-GDRG-----RGGRRRAGGR 42
GG RGG G GGRGG RGG GGR GDRG RGG R GGR
Sbjct: 87 GGFRGGRGGDRGGRGGGRGGFGGRGGDRGTPFKARGGGRGGGGR 218
Score = 31.6 bits (70), Expect = 0.35
Identities = 20/33 (60%), Positives = 20/33 (60%), Gaps = 1/33 (3%)
Frame = +3
Query: 12 GSGFGGRGGDRGG-RGGRGDRGRGGRRRAGGRR 43
G G GG G RGG RGGRG GRGG GG R
Sbjct: 75 GRGGGGFRGGRGGDRGGRGG-GRGGFGGRGGDR 170
>TC78850 homologue to GP|10177413|dbj|BAB10544. fibrillarin-like
{Arabidopsis thaliana}, partial (87%)
Length = 1177
Score = 48.1 bits (113), Expect = 4e-06
Identities = 28/48 (58%), Positives = 29/48 (60%), Gaps = 8/48 (16%)
Frame = +3
Query: 3 ERGGDRGGFGSGFGG--------RGGDRGGRGGRGDRGRGGRRRAGGR 42
+RGG GG G GFGG RGG RG GGRG RG GGR R GGR
Sbjct: 93 DRGGRGGGGGRGFGGGRGGDFKPRGGGRGFGGGRGARG-GGRGRGGGR 233
Score = 47.8 bits (112), Expect = 5e-06
Identities = 24/35 (68%), Positives = 24/35 (68%), Gaps = 6/35 (17%)
Frame = +3
Query: 14 GFGGRGGDRGGRGGRGDRGRGGRR------RAGGR 42
GFGGRGGDRGGRGG G RG GG R R GGR
Sbjct: 72 GFGGRGGDRGGRGGGGGRGFGGGRGGDFKPRGGGR 176
Score = 37.0 bits (84), Expect = 0.008
Identities = 19/33 (57%), Positives = 20/33 (60%)
Frame = +3
Query: 4 RGGDRGGFGSGFGGRGGDRGGRGGRGDRGRGGR 36
RGG RG FG G G RGG RG GGRG G +
Sbjct: 162 RGGGRG-FGGGRGARGGGRGRGGGRGGMKGGSK 257
>BQ750872 similar to PIR|A36363|R3BY ribosomal protein S2.e cytosolic -
yeast (Saccharomyces cerevisiae), partial (12%)
Length = 364
Score = 47.0 bits (110), Expect = 8e-06
Identities = 19/37 (51%), Positives = 25/37 (67%)
Frame = -3
Query: 226 VKATFDCLMKTYGFLTPEFWKETRFSKSPFQEYTDLL 262
+KAT + TYGFLTP WKET+ +SP +E+ D L
Sbjct: 359 LKATLVAVSNTYGFLTPNLWKETKLIRSPLEEFADTL 249
>TC79934 similar to GP|20259633|gb|AAM14173.1 putative GAR1 protein
{Arabidopsis thaliana}, partial (87%)
Length = 814
Score = 45.4 bits (106), Expect = 2e-05
Identities = 28/41 (68%), Positives = 28/41 (68%), Gaps = 2/41 (4%)
Frame = +1
Query: 4 RGGDRGGFGSGF-GGR-GGDRGGRGGRGDRGRGGRRRAGGR 42
RGG RGG GF GGR GG RGGR G G RGRGG R GGR
Sbjct: 67 RGGGRGG---GFRGGRDGGFRGGRDGGGFRGRGGGRFGGGR 180
Score = 37.7 bits (86), Expect = 0.005
Identities = 25/49 (51%), Positives = 25/49 (51%), Gaps = 11/49 (22%)
Frame = +1
Query: 5 GGDRGGFGSGFGGRGGDRGG---------RGGRGD--RGRGGRRRAGGR 42
GG GG G G GRGG RGG RGGRG GRGG R GR
Sbjct: 508 GGRGGGRGGGGFGRGGGRGGGGFRGRGPPRGGRGPPRGGRGGGFRGRGR 654
Score = 37.0 bits (84), Expect = 0.008
Identities = 24/45 (53%), Positives = 24/45 (53%), Gaps = 10/45 (22%)
Frame = +1
Query: 4 RGGDRGG---------FGSGFGGRGGDRGGRG-GRGDRGRGGRRR 38
RGG RGG G GF GRG RGGRG RG RG G R R
Sbjct: 514 RGGGRGGGGFGRGGGRGGGGFRGRGPPRGGRGPPRGGRGGGFRGR 648
Score = 33.5 bits (75), Expect = 0.093
Identities = 21/39 (53%), Positives = 21/39 (53%), Gaps = 8/39 (20%)
Frame = +1
Query: 12 GSGFGGRGGDR--------GGRGGRGDRGRGGRRRAGGR 42
G GGRGG R GGRGG G RGRG R GGR
Sbjct: 496 GQASGGRGGGRGGGGFGRGGGRGGGGFRGRGPPR--GGR 606
Score = 28.5 bits (62), Expect = 3.0
Identities = 14/23 (60%), Positives = 14/23 (60%)
Frame = +1
Query: 4 RGGDRGGFGSGFGGRGGDRGGRG 26
RG RGG G GGRGG GRG
Sbjct: 583 RGPPRGGRGPPRGGRGGGFRGRG 651
>AL385184 weakly similar to SP|Q90YS3|RS2_I 40S ribosomal protein S2.
[Channel catfish] {Ictalurus punctatus}, partial (10%)
Length = 203
Score = 44.7 bits (104), Expect = 4e-05
Identities = 18/33 (54%), Positives = 24/33 (72%)
Frame = +3
Query: 236 TYGFLTPEFWKETRFSKSPFQEYTDLLAKPTAK 268
TYGFLTP+ WK+ SP+QE++D+LAK K
Sbjct: 6 TYGFLTPDLWKDGDIKLSPYQEFSDMLAKAPDK 104
>BQ141909 weakly similar to GP|14253159|emb VMP3 protein {Volvox carteri f.
nagariensis}, partial (13%)
Length = 1223
Score = 44.3 bits (103), Expect = 5e-05
Identities = 24/43 (55%), Positives = 26/43 (59%), Gaps = 3/43 (6%)
Frame = +2
Query: 2 AERGGDRGGFGSGFGGRGGDRGGR---GGRGDRGRGGRRRAGG 41
AE G +RG G GGRGG R GR GG G +GRG R R GG
Sbjct: 215 AEGGSERGRGRRGGGGRGGGRRGRGKKGGGGTKGRGKRERGGG 343
Score = 40.8 bits (94), Expect = 6e-04
Identities = 24/46 (52%), Positives = 26/46 (56%), Gaps = 3/46 (6%)
Frame = +2
Query: 1 MAERG--GDRGGFGSGFGG-RGGDRGGRGGRGDRGRGGRRRAGGRR 43
M RG G RG G G GG GG GRG RG GRGG RR G++
Sbjct: 158 MRRRGERGKRGEGGEGGGGAEGGSERGRGRRGGGGRGGGRRGRGKK 295
Score = 38.5 bits (88), Expect = 0.003
Identities = 24/46 (52%), Positives = 26/46 (56%), Gaps = 1/46 (2%)
Frame = +2
Query: 4 RGGDRGGFGSGFGGRGGDRGG-RGGRGDRGRGGRRRAGGRREEEEK 48
R G+RG G G G GG GG GRG RG GGR GGRR +K
Sbjct: 164 RRGERGKRGEGGEGGGGAEGGSERGRGRRGGGGR--GGGRRGRGKK 295
Score = 38.5 bits (88), Expect = 0.003
Identities = 27/60 (45%), Positives = 31/60 (51%)
Frame = +3
Query: 6 GDRGGFGSGFGGRGGDRGGRGGRGDRGRGGRRRAGGRREEEEKWVPVTKLGRLVKEGKIR 65
G RGG G G RGG RG GG G G G GGR+ EE+ K G V+EG+ R
Sbjct: 186 GGRGGRGGG-ERRGGRRGEEGGGGGGGGEGGGGGGGRKGGEEQ-----KEGGSVREGEER 347
Score = 38.1 bits (87), Expect = 0.004
Identities = 21/45 (46%), Positives = 25/45 (54%)
Frame = +3
Query: 4 RGGDRGGFGSGFGGRGGDRGGRGGRGDRGRGGRRRAGGRREEEEK 48
RGG RG G GG GG GG GG G +G ++ G RE EE+
Sbjct: 219 RGGRRG--EEGGGGGGGGEGGGGGGGRKGGEEQKEGGSVREGEER 347
Score = 34.7 bits (78), Expect = 0.042
Identities = 19/37 (51%), Positives = 21/37 (56%)
Frame = +2
Query: 5 GGDRGGFGSGFGGRGGDRGGRGGRGDRGRGGRRRAGG 41
GG+ GG G RG R G GGRG GR GR + GG
Sbjct: 194 GGEGGGGAEGGSERGRGRRGGGGRGG-GRRGRGKKGG 301
Score = 34.3 bits (77), Expect = 0.054
Identities = 23/44 (52%), Positives = 25/44 (56%), Gaps = 4/44 (9%)
Frame = +2
Query: 5 GGDRGGFGSGFGGRGGDRGGRGGRGDRGR----GGRRRAGGRRE 44
GG GG G G RGG GGRGG G RGR GG + G+RE
Sbjct: 209 GGAEGGSERGRGRRGG--GGRGG-GRRGRGKKGGGGTKGRGKRE 331
Score = 33.9 bits (76), Expect = 0.071
Identities = 22/48 (45%), Positives = 25/48 (51%), Gaps = 4/48 (8%)
Frame = +1
Query: 3 ERGGDRGGFGSGFGGRGGDRGGRGGRG----DRGRGGRRRAGGRREEE 46
E GG GG G G G G +RGGR R +RGR G R G R +E
Sbjct: 244 EEGG--GGEGRGEEGEGEERGGRNKRKGEA*ERGRNGGGRQGKNRGKE 381
Score = 28.9 bits (63), Expect = 2.3
Identities = 16/31 (51%), Positives = 17/31 (54%), Gaps = 4/31 (12%)
Frame = +2
Query: 16 GGRGGDRGGRGGRGDRGRGG----RRRAGGR 42
GGR GDRGG G R R RR+ GGR
Sbjct: 563 GGRAGDRGGGGLRAPSKRRNSKTKRRKRGGR 655
Score = 28.9 bits (63), Expect = 2.3
Identities = 23/71 (32%), Positives = 29/71 (40%), Gaps = 18/71 (25%)
Frame = +2
Query: 5 GGDRGGFGSGFGGRGGDRGGR----------------GGRGDRGRGGRRRA--GGRREEE 46
GG+R G G R +R R GGR R GRRRA GGR E+
Sbjct: 1010 GGERTGESGGGE*RQSERERRRERRRTRKQIAEEKK*GGREGRSETGRRRAGGGGREREQ 1189
Query: 47 EKWVPVTKLGR 57
+ + + GR
Sbjct: 1190 RREMKARRRGR 1222
Score = 27.3 bits (59), Expect = 6.6
Identities = 21/59 (35%), Positives = 28/59 (46%), Gaps = 4/59 (6%)
Frame = +1
Query: 5 GGDRGGFGSGFGG---RGGDRGGRGGRGDRGRGGRRRAGGR-REEEEKWVPVTKLGRLV 59
G +RGG G RG + GGR G+ R AGG+ R + E+ +T GR V
Sbjct: 286 GEERGGRNKRKGEA*ERGRNGGGRQGKNRGKEWDREEAGGKWRGKREREKRITTNGRRV 462
Score = 26.9 bits (58), Expect = 8.7
Identities = 22/59 (37%), Positives = 27/59 (45%), Gaps = 14/59 (23%)
Frame = +2
Query: 4 RGGDRGGFGSGFGG---------RGG--DRGGRGGRGDRGRGGRRR---AGGRREEEEK 48
RGG R G G GG RGG + GG+G R GRRR GG+ E+ +
Sbjct: 263 RGGGRRGRGKKGGGGTKGRGKRERGGGTEGGGKGKIEGRNGTGRRRGESGGGKGREKSE 439
>TC88887 weakly similar to GP|6523547|emb|CAB62280.1 hydroxyproline-rich
glycoprotein DZ-HRGP {Volvox carteri f. nagariensis},
partial (18%)
Length = 1681
Score = 43.9 bits (102), Expect = 7e-05
Identities = 24/38 (63%), Positives = 25/38 (65%), Gaps = 1/38 (2%)
Frame = -3
Query: 5 GGDRGGFGSGFGGRGGDRGGRGG-RGDRGRGGRRRAGG 41
GG RGGFG G+ GRGG GGRGG G GRGG GG
Sbjct: 689 GGMRGGFGGGY-GRGGFNGGRGGFGGGYGRGGFAGGGG 579
Score = 32.3 bits (72), Expect = 0.21
Identities = 15/22 (68%), Positives = 16/22 (72%)
Frame = -3
Query: 6 GDRGGFGSGFGGRGGDRGGRGG 27
G RGGFG G+ GRGG GG GG
Sbjct: 638 GGRGGFGGGY-GRGGFAGGGGG 576
Score = 31.2 bits (69), Expect = 0.46
Identities = 19/30 (63%), Positives = 19/30 (63%), Gaps = 1/30 (3%)
Frame = -3
Query: 14 GFGGRGGDRGGRGG-RGDRGRGGRRRAGGR 42
GFGGRGG G RGG G GRGG GGR
Sbjct: 713 GFGGRGGFGGMRGGFGGGYGRGGFN--GGR 630
Score = 28.9 bits (63), Expect = 2.3
Identities = 21/44 (47%), Positives = 22/44 (49%), Gaps = 3/44 (6%)
Frame = -3
Query: 2 AERGGDRG-GFGSGFGGRGGDRGGRGGRG--DRGRGGRRRAGGR 42
A GG G G GFGG G GG GRG + GRGG GR
Sbjct: 734 AGAGGPMGFGGRGGFGGMRGGFGGGYGRGGFNGGRGGFGGGYGR 603
>TC76523 homologue to PIR|T09648|T09648 nucleolin homolog nuM1 - alfalfa,
partial (69%)
Length = 1615
Score = 43.5 bits (101), Expect = 9e-05
Identities = 23/38 (60%), Positives = 23/38 (60%), Gaps = 1/38 (2%)
Frame = +3
Query: 5 GGDRGGFGSGFGGRGGDR-GGRGGRGDRGRGGRRRAGG 41
GG GGFG GGRGG R GGR G GRGGR GG
Sbjct: 1161 GGRSGGFGGRSGGRGGGRFGGRDSGGRGGRGGRGGRGG 1274
Score = 39.7 bits (91), Expect = 0.001
Identities = 25/56 (44%), Positives = 27/56 (47%), Gaps = 15/56 (26%)
Frame = +3
Query: 2 AERGGDRGGFGS-------------GFGGRGGDRGGR--GGRGDRGRGGRRRAGGR 42
++ G RGG G GFGGR G RGG GGR GRGGR GGR
Sbjct: 1101 SQGSGGRGGGGRSGGGRFGGGGRSGGFGGRSGGRGGGRFGGRDSGGRGGRGGRGGR 1268
>TC89001 similar to GP|5726567|gb|AAD48471.1| glycine-rich RNA-binding
protein {Glycine max}, partial (90%)
Length = 488
Score = 43.5 bits (101), Expect = 9e-05
Identities = 23/39 (58%), Positives = 24/39 (60%)
Frame = +3
Query: 5 GGDRGGFGSGFGGRGGDRGGRGGRGDRGRGGRRRAGGRR 43
GG RGG G G+GG GG GG GG G G GGRR G R
Sbjct: 300 GGGRGGGGGGYGGGGGYGGGGGGYG--GGGGRRDGGYSR 410
Score = 43.5 bits (101), Expect = 9e-05
Identities = 22/38 (57%), Positives = 24/38 (62%)
Frame = +3
Query: 4 RGGDRGGFGSGFGGRGGDRGGRGGRGDRGRGGRRRAGG 41
RGG GG+G G GG GG GG GG G R GG R+GG
Sbjct: 309 RGGGGGGYGGG-GGYGGGGGGYGGGGGRRDGGYSRSGG 419
Score = 37.0 bits (84), Expect = 0.008
Identities = 18/30 (60%), Positives = 18/30 (60%)
Frame = +3
Query: 12 GSGFGGRGGDRGGRGGRGDRGRGGRRRAGG 41
GSG GGRGG GG GG G G GG GG
Sbjct: 291 GSGGGGRGGGGGGYGGGGGYGGGGGGYGGG 380
>TC85851 similar to PIR|T12180|T12180 probable transcription factor - fava
bean, partial (93%)
Length = 2007
Score = 43.5 bits (101), Expect = 9e-05
Identities = 22/31 (70%), Positives = 22/31 (70%)
Frame = +3
Query: 4 RGGDRGGFGSGFGGRGGDRGGRGGRGDRGRG 34
RGGDR GFGGRGGDRG GGRG RG G
Sbjct: 465 RGGDRD---RGFGGRGGDRGFGGGRGGRGFG 548
Score = 40.8 bits (94), Expect = 6e-04
Identities = 27/41 (65%), Positives = 27/41 (65%), Gaps = 4/41 (9%)
Frame = +3
Query: 6 GDRGGFGSGFGGRGGDR----GGRGGRGDRGRGGRRRAGGR 42
G RGG GFGGRGGDR GGRG GDRG GG R GGR
Sbjct: 435 GGRGG--RGFGGRGGDRDRGFGGRG--GDRGFGGGR--GGR 539
Score = 29.6 bits (65), Expect = 1.3
Identities = 13/20 (65%), Positives = 14/20 (70%)
Frame = +3
Query: 21 DRGGRGGRGDRGRGGRRRAG 40
+ GGRGGRG GRGG R G
Sbjct: 429 NEGGRGGRGFGGRGGDRDRG 488
Score = 29.3 bits (64), Expect = 1.7
Identities = 13/19 (68%), Positives = 13/19 (68%)
Frame = +3
Query: 16 GGRGGDRGGRGGRGDRGRG 34
GGRG RGGRG RG RG
Sbjct: 1236 GGRGRGRGGRGARGGGFRG 1292
Score = 28.1 bits (61), Expect = 3.9
Identities = 14/19 (73%), Positives = 14/19 (73%)
Frame = +3
Query: 4 RGGDRGGFGSGFGGRGGDR 22
RGGDRG FG G GGRG R
Sbjct: 498 RGGDRG-FGGGRGGRGFGR 551
Score = 27.3 bits (59), Expect = 6.6
Identities = 20/51 (39%), Positives = 23/51 (44%), Gaps = 7/51 (13%)
Frame = +3
Query: 4 RGGDRGGFGSGFGGRGG------DRGGRGGRG-DRGRGGRRRAGGRREEEE 47
RGG RGGF +G G G +R GRG D R G R E +E
Sbjct: 672 RGGRRGGFNNGDAGEEGRPRRTFERHSGTGRGNDFKREGAGRGNWGTETDE 824
>BQ144407 weakly similar to GP|6523547|emb| hydroxyproline-rich glycoprotein
DZ-HRGP {Volvox carteri f. nagariensis}, partial (23%)
Length = 1217
Score = 43.1 bits (100), Expect = 1e-04
Identities = 29/70 (41%), Positives = 32/70 (45%), Gaps = 17/70 (24%)
Frame = +1
Query: 5 GGDRGGFGSGFGGRGGDRGGRGGRGDRGRGGRR-----------------RAGGRREEEE 47
GG G G G+GGRG GG GGRG G GGRR R GG R EEE
Sbjct: 136 GGGINGNGEGWGGRGRPGGGGGGRG--GGGGRRNGGEIGALDREQR*GDDRGGGMRMEEE 309
Query: 48 KWVPVTKLGR 57
P+ + R
Sbjct: 310 VGEPIRGMKR 339
Score = 33.1 bits (74), Expect = 0.12
Identities = 28/72 (38%), Positives = 33/72 (44%), Gaps = 17/72 (23%)
Frame = +3
Query: 3 ERGGDRG-----------GFGSGFGGRGG--DRGGRGGRGDRGRGGRRR----AGGRREE 45
+RGGDRG G+ GGRGG D+G G G R GGRR GG E
Sbjct: 228 KRGGDRGLGPGAAIGG*PRGGNADGGRGGGADKGHEAG-GQRNGGGRREWAAGRGGGVGE 404
Query: 46 EEKWVPVTKLGR 57
E V ++GR
Sbjct: 405 ESAVQRVKRMGR 440
Score = 31.2 bits (69), Expect = 0.46
Identities = 26/73 (35%), Positives = 34/73 (45%), Gaps = 10/73 (13%)
Frame = +1
Query: 6 GDRGGF---GSGFGGRGGDRG-------GRGGRGDRGRGGRRRAGGRREEEEKWVPVTKL 55
GDR G+ G G GGR G G GRGG G RGGRR+ E +W ++
Sbjct: 541 GDRRGW*GGGRGEGGRTGGWGSDRDVERGRGGYG--VRGGRRKG---VNESRRWGRRVQV 705
Query: 56 GRLVKEGKIRSLE 68
+E + R +E
Sbjct: 706 RDAAREAREREME 744
>TC76561 similar to PIR|S71779|S71779 glycine-rich RNA-binding protein GRP1
- wheat, partial (95%)
Length = 1054
Score = 40.8 bits (94), Expect = 6e-04
Identities = 23/43 (53%), Positives = 25/43 (57%), Gaps = 5/43 (11%)
Frame = +1
Query: 4 RGGDRGGFGSGFGGRGGDRGGRGGRGDRGRG-----GRRRAGG 41
RGG GG+G G GG GG+R G GG G G G G RR GG
Sbjct: 622 RGG--GGYGGGGGGYGGERRGYGGGGGYGGGGGGGYGERRGGG 744
Score = 37.4 bits (85), Expect = 0.006
Identities = 22/39 (56%), Positives = 25/39 (63%), Gaps = 2/39 (5%)
Frame = +1
Query: 5 GGDRGGFGSGFGGRGGDRGGRGGRGDR--GRGGRRRAGG 41
GG+R G+G G GG GG GG GG G+R G GG R GG
Sbjct: 661 GGERRGYGGG-GGYGG--GGGGGYGERRGGGGGYSRGGG 768
Score = 36.6 bits (83), Expect = 0.011
Identities = 18/36 (50%), Positives = 20/36 (55%)
Frame = +1
Query: 6 GDRGGFGSGFGGRGGDRGGRGGRGDRGRGGRRRAGG 41
G GG+G G GG G+R G GG RG GG GG
Sbjct: 682 GGGGGYGGGGGGGYGERRGGGGGYSRGGGGGGYGGG 789
Score = 29.6 bits (65), Expect = 1.3
Identities = 13/23 (56%), Positives = 14/23 (60%)
Frame = +1
Query: 5 GGDRGGFGSGFGGRGGDRGGRGG 27
G RGG G G+GG G RGG G
Sbjct: 748 GYSRGGGGGGYGGGGYSRGGGDG 816
Score = 28.1 bits (61), Expect = 3.9
Identities = 12/21 (57%), Positives = 13/21 (61%)
Frame = +1
Query: 4 RGGDRGGFGSGFGGRGGDRGG 24
RGG GG+G G RGG GG
Sbjct: 757 RGGGGGGYGGGGYSRGGGDGG 819
>TC76556 similar to PIR|S71779|S71779 glycine-rich RNA-binding protein GRP1
- wheat, partial (95%)
Length = 754
Score = 40.8 bits (94), Expect = 6e-04
Identities = 23/43 (53%), Positives = 25/43 (57%), Gaps = 5/43 (11%)
Frame = +2
Query: 4 RGGDRGGFGSGFGGRGGDRGGRGGRGDRGRG-----GRRRAGG 41
RGG GG+G G GG GG+R G GG G G G G RR GG
Sbjct: 368 RGG--GGYGGGGGGYGGERRGYGGGGGYGGGGGGGYGERRGGG 490
Score = 37.4 bits (85), Expect = 0.006
Identities = 22/39 (56%), Positives = 25/39 (63%), Gaps = 2/39 (5%)
Frame = +2
Query: 5 GGDRGGFGSGFGGRGGDRGGRGGRGDR--GRGGRRRAGG 41
GG+R G+G G GG GG GG GG G+R G GG R GG
Sbjct: 407 GGERRGYGGG-GGYGG--GGGGGYGERRGGGGGYSRGGG 514
Score = 36.6 bits (83), Expect = 0.011
Identities = 18/36 (50%), Positives = 20/36 (55%)
Frame = +2
Query: 6 GDRGGFGSGFGGRGGDRGGRGGRGDRGRGGRRRAGG 41
G GG+G G GG G+R G GG RG GG GG
Sbjct: 428 GGGGGYGGGGGGGYGERRGGGGGYSRGGGGGGYGGG 535
Score = 29.6 bits (65), Expect = 1.3
Identities = 13/23 (56%), Positives = 14/23 (60%)
Frame = +2
Query: 5 GGDRGGFGSGFGGRGGDRGGRGG 27
G RGG G G+GG G RGG G
Sbjct: 494 GYSRGGGGGGYGGGGYSRGGGDG 562
Score = 28.1 bits (61), Expect = 3.9
Identities = 12/21 (57%), Positives = 13/21 (61%)
Frame = +2
Query: 4 RGGDRGGFGSGFGGRGGDRGG 24
RGG GG+G G RGG GG
Sbjct: 503 RGGGGGGYGGGGYSRGGGDGG 565
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.319 0.140 0.418
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 7,539,397
Number of Sequences: 36976
Number of extensions: 101981
Number of successful extensions: 2368
Number of sequences better than 10.0: 246
Number of HSP's better than 10.0 without gapping: 1353
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1988
length of query: 279
length of database: 9,014,727
effective HSP length: 95
effective length of query: 184
effective length of database: 5,502,007
effective search space: 1012369288
effective search space used: 1012369288
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 58 (26.9 bits)
Medicago: description of AC140104.12


