

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC139344.2 - phase: 0
(1591 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
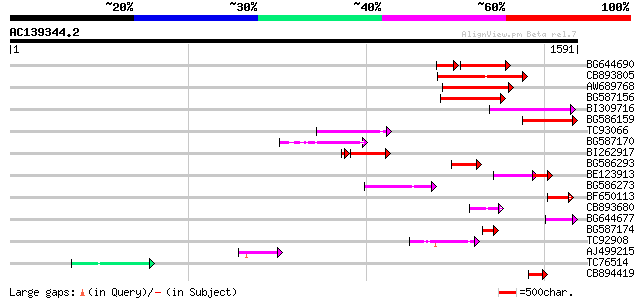
Score E
Sequences producing significant alignments: (bits) Value
BG644690 weakly similar to GP|18542179|gb putative pol protein {... 194 3e-66
CB893805 similar to GP|10177935|d copia-type polyprotein {Arabid... 179 1e-44
AW689768 weakly similar to GP|10177485|d polyprotein {Arabidopsi... 172 7e-43
BG587156 similar to PIR|G85055|G8 probable polyprotein [imported... 165 1e-40
BI309716 weakly similar to PIR|G96722|G9 hypothetical protein F2... 159 8e-39
BG586159 weakly similar to PIR|T47841|T4 hypothetical protein T2... 139 1e-32
TC93066 weakly similar to GP|19920130|gb|AAM08562.1 Putative ret... 124 4e-28
BG587170 similar to PIR|F86470|F8 probable retroelement polyprot... 117 4e-26
BI262917 weakly similar to GP|19920130|g Putative retroelement {... 102 2e-22
BG586293 weakly similar to PIR|E84473|E84 probable retroelement ... 84 3e-16
BE123913 weakly similar to GP|22093573|d polyprotein {Oryza sati... 81 3e-15
BG586273 weakly similar to PIR|F86470|F8 probable retroelement p... 80 5e-15
BF650113 weakly similar to GP|4753889|emb| Tpv2-1c {Phaseolus vu... 68 3e-11
CB893680 weakly similar to GP|1167523|db ORF(AA 1-1338) {Nicotia... 62 1e-09
BG644677 weakly similar to GP|7682800|gb| Hypothetical protein T... 58 3e-08
BG587174 similar to PIR|A47759|A4775 retrovirus-related reverse ... 58 3e-08
TC92908 similar to GP|10140704|gb|AAG13538.1 putative gag-pol po... 55 2e-07
AJ499215 weakly similar to GP|6642775|gb| gag-pol polyprotein {V... 54 6e-07
TC76514 homologue to PIR|T09648|T09648 nucleolin homolog nuM1 - ... 50 7e-06
CB894419 weakly similar to GP|10177935|db copia-type polyprotein... 50 7e-06
>BG644690 weakly similar to GP|18542179|gb putative pol protein {Zea mays},
partial (22%)
Length = 629
Score = 194 bits (492), Expect(2) = 3e-66
Identities = 88/141 (62%), Positives = 119/141 (83%)
Frame = -2
Query: 1265 VTRNKARLVAQGYSQQEGINYTETFAPVARLETIRLLLSYAINHGIILYQMDVKSVFLNG 1324
+TRNK++LV QGY+Q+EGI+Y E F+PVAR+E IR+L+++A G LYQMDVKS F+NG
Sbjct: 424 ITRNKSKLVVQGYNQKEGIDYDEAFSPVARMEAIRILIAFAAFMGFKLYQMDVKSAFING 245
Query: 1325 VIEEEVYVKQPPGFEDLKHPDHVYKLKKSLYGLKQAPRAWYDRLSNFLIKNDFERGQVDT 1384
++EEV+VKQPPGFED + P+HV++L K+LYGLKQAPRAWY+RLS FL+KN F+RG++D
Sbjct: 244 DLKEEVFVKQPPGFEDAEVPNHVFRLNKTLYGLKQAPRAWYERLSKFLLKNGFKRGKIDN 65
Query: 1385 TLFRRTLKKDILIVQIYVDDI 1405
TLF + ++LI+Q+YVDDI
Sbjct: 64 TLFLLKRE*ELLIIQVYVDDI 2
Score = 78.6 bits (192), Expect(2) = 3e-66
Identities = 37/62 (59%), Positives = 46/62 (73%)
Frame = -3
Query: 1198 SLIGLLSIIEPKTVEEALSDDGWILAMQEELNQFQRNDVWDLVPKPFQKNIIETKWVFRN 1257
S +S IEPK V+EAL D WI +MQEEL+QF+R+ VW LVP+P K +I T+WVFRN
Sbjct: 615 SFSAFISSIEPKNVKEALRDADWINSMQEELHQFERSKVWYLVPRPEGKTVIGTRWVFRN 436
Query: 1258 KL 1259
KL
Sbjct: 435 KL 430
>CB893805 similar to GP|10177935|d copia-type polyprotein {Arabidopsis
thaliana}, partial (14%)
Length = 778
Score = 179 bits (453), Expect = 1e-44
Identities = 95/257 (36%), Positives = 157/257 (60%), Gaps = 5/257 (1%)
Frame = +3
Query: 1200 IGLLSII-EPKTVEEALSDDGWILAMQEELNQFQRNDVWDLVPKPFQKNIIETKWVFRNK 1258
+G+L++ +P T EEA+ + W +M E+ +RN+ W+L I KW+F+ K
Sbjct: 15 LGMLTMTSDPTTFEEAVKSEKWRASMNNEMEATERNNTWELTDLRSGAKTIGLKWIFKTK 194
Query: 1259 LNEQGEVTRNKARLVAQGYSQQEGINYTETFAPVARLETIRLLLSYAI---NHGIILYQM 1315
LNE GE+ + KARLVA+GYSQQ G++YTE FAPVAR +TIR++++ A G+ + M
Sbjct: 195 LNENGEIEKYKARLVAKGYSQQYGVDYTEVFAPVARWDTIRMVIALAAQIKRDGVCIS*M 374
Query: 1316 DVKSVFLNGVIEEEVYVKQPPGFEDLKHPDHVYKLKKSLYGLKQAPRAWYDRLSNFLIKN 1375
+ + + + + + ++K++LYGLKQAPRAWY R+ + K
Sbjct: 375 *KAHSCMEN*MRKFLLINH----RVM*RRVIS*RVKRALYGLKQAPRAWYSRIEAYFTKE 542
Query: 1376 DFERGQVDTTLFRRTLK-KDILIVQIYVDDIIFGSTNASLCKEFSKLMQDEFEMSMMGEL 1434
FE+ + TLF + + ILI+ +YVDD+IF + ++ +EF K M+ EF MS +G++
Sbjct: 543 GFEKCPYEHTLFVKLSEGGKILIISLYVDDLIFIGNDENMFEEFKKSMKKEFNMSDLGKM 722
Query: 1435 KFFLGIQINQSKEGVYV 1451
+FLG+++ Q+++G+Y+
Sbjct: 723 HYFLGVEVTQNEKGIYI 773
>AW689768 weakly similar to GP|10177485|d polyprotein {Arabidopsis thaliana},
partial (9%)
Length = 675
Score = 172 bits (437), Expect = 7e-43
Identities = 93/199 (46%), Positives = 125/199 (62%)
Frame = +1
Query: 1215 LSDDGWILAMQEELNQFQRNDVWDLVPKPFQKNIIETKWVFRNKLNEQGEVTRNKARLVA 1274
LSD W+ AM+ E N WDLVP P K I KWV+R K N G V + KARLVA
Sbjct: 82 LSDPRWLQAMKTEYKALIDNKTWDLVPLPPHKKAIGCKWVYRVKENPDGSVNKFKARLVA 261
Query: 1275 QGYSQQEGINYTETFAPVARLETIRLLLSYAINHGIILYQMDVKSVFLNGVIEEEVYVKQ 1334
+G+SQ G +YTETF+PV + TIRL+L+ AI + + Q+D+ + FLNG ++EEVY+ Q
Sbjct: 262 KGFSQTLGCDYTETFSPVIKPVTIRLILTIAITYKWEIQQIDINNAFLNGFLQEEVYMSQ 441
Query: 1335 PPGFEDLKHPDHVYKLKKSLYGLKQAPRAWYDRLSNFLIKNDFERGQVDTTLFRRTLKKD 1394
P GFE + V KL KSLYGLKQAPRAWY+ L++ I+ F + + D +L
Sbjct: 442 PQGFE-AANKSLVCKLNKSLYGLKQAPRAWYEXLTSAQIQFGFTKSRCDPSLLIYNQNGA 618
Query: 1395 ILIVQIYVDDIIFGSTNAS 1413
+ + IYVDDI+ ++AS
Sbjct: 619 CIYLXIYVDDILITGSSAS 675
>BG587156 similar to PIR|G85055|G8 probable polyprotein [imported] -
Arabidopsis thaliana, partial (17%)
Length = 618
Score = 165 bits (417), Expect = 1e-40
Identities = 82/183 (44%), Positives = 116/183 (62%)
Frame = -1
Query: 1208 PKTVEEALSDDGWILAMQEELNQFQRNDVWDLVPKPFQKNIIETKWVFRNKLNEQGEVTR 1267
P++ EEA+ D W ++ E +ND W P K + ++W+F K G + R
Sbjct: 558 PRSYEEAMEDKEWKESVGAEAGAMIKNDTWYESELPKGKKAVSSRWIFTIKYKADGSIER 379
Query: 1268 NKARLVAQGYSQQEGINYTETFAPVARLETIRLLLSYAINHGIILYQMDVKSVFLNGVIE 1327
K RLVA+G++ G +Y ETFAPVA+L TIR++LS A+N G L+QMDVK+ FL G +E
Sbjct: 378 KKTRLVARGFTLTYGEDYIETFAPVAKLHTIRIVLSLAVNLGWGLWQMDVKNAFLQGELE 199
Query: 1328 EEVYVKQPPGFEDLKHPDHVYKLKKSLYGLKQAPRAWYDRLSNFLIKNDFERGQVDTTLF 1387
+EVY+ PPG E L +V +LKK++YGLKQ+PRAWY++LS L F + ++D TLF
Sbjct: 198 DEVYMYPPPGLEHLVKRGNVLRLKKAIYGLKQSPRAWYNKLSTTLNGRGFRKSELDHTLF 19
Query: 1388 RRT 1390
T
Sbjct: 18 TLT 10
>BI309716 weakly similar to PIR|G96722|G9 hypothetical protein F20P5.25
[imported] - Arabidopsis thaliana, partial (10%)
Length = 744
Score = 159 bits (402), Expect = 8e-39
Identities = 88/240 (36%), Positives = 134/240 (55%)
Frame = +2
Query: 1347 VYKLKKSLYGLKQAPRAWYDRLSNFLIKNDFERGQVDTTLFRRTLKKDILIVQIYVDDII 1406
V +L+KS+YGLKQA R WY +LS LI + + D +LF + + +YVDDI+
Sbjct: 20 VCELQKSIYGLKQASRQWYSKLSESLISFGYLQSSSDFSLFTKFKDSSFTTLLVYVDDIV 199
Query: 1407 FGSTNASLCKEFSKLMQDEFEMSMMGELKFFLGIQINQSKEGVYVHQTKYTKELLKKFKL 1466
+ S + + D F++ +G L++FLG+++ +SK+G+ ++Q KYT ELL+
Sbjct: 200 LAGNDISEIQHVKCFLIDRFKIKDLGSLRYFLGLEVARSKQGILLNQRKYTLELLEDSGN 379
Query: 1467 EDCKVMNTPMHPTCTLSKEDTGTVVDQKLYRGMIGSLLYLTASRPDILFSVCLCARFQSD 1526
K TP + L D+ D+ YR +IG L+YLT +RPDI F+V ++F S
Sbjct: 380 LAVKSTLTPYDISLKLHNSDSPLYNDETQYRRLIGKLIYLTTTRPDISFAVQQLSQFVSK 559
Query: 1527 PRESHLTAVKRIFRYLKGTTNLGLLYRKSLDYKLIGFCDADYAGDRIERKSTSGNCQFLG 1586
P++ H A R+ +YLK GL Y + + KL F D+D+A RKS +G FLG
Sbjct: 560 PQQVHYQAAIRVLQYLKTAPAKGLFYSATSNLKLSSFADSDWATCPTTRKSVTGYWVFLG 739
>BG586159 weakly similar to PIR|T47841|T4 hypothetical protein T2O9.150 -
Arabidopsis thaliana, partial (11%)
Length = 732
Score = 139 bits (349), Expect = 1e-32
Identities = 63/152 (41%), Positives = 100/152 (65%)
Frame = +1
Query: 1440 IQINQSKEGVYVHQTKYTKELLKKFKLEDCKVMNTPMHPTCTLSKEDTGTVVDQKLYRGM 1499
+++ Q++EG+Y+ Q KY +LL++F +E + P+ P C L K++ G VD Y+ +
Sbjct: 1 VEVIQNEEGIYICQRKYVTDLLERFGMEKSNLSRNPIAPRCKLIKDENGVKVDATKYKQI 180
Query: 1500 IGSLLYLTASRPDILFSVCLCARFQSDPRESHLTAVKRIFRYLKGTTNLGLLYRKSLDYK 1559
+G L+YL A+RPD+++ + L +RF + P E H+ AVKR+ RYL GT NLG++Y+++ K
Sbjct: 181 VGCLMYLAATRPDLMYVLSLISRFMNCPTELHMHAVKRVLRYLNGTINLGIMYKRNGSEK 360
Query: 1560 LIGFCDADYAGDRIERKSTSGNCQFLGENLIS 1591
L + D+DYAGD +RKSTSG L +S
Sbjct: 361 LEAYTDSDYAGDLDDRKSTSGYVFMLSSGAVS 456
>TC93066 weakly similar to GP|19920130|gb|AAM08562.1 Putative retroelement
{Oryza sativa} [Oryza sativa (japonica cultivar-group)],
partial (10%)
Length = 823
Score = 124 bits (310), Expect = 4e-28
Identities = 72/214 (33%), Positives = 109/214 (50%), Gaps = 2/214 (0%)
Frame = +1
Query: 860 LELLHIDLFGPVNTASLYGSKYGLVIVDDYSRWTWVKFIKSKDYACEVFSSFCTQIQSEK 919
L+ +H DL+GP S G +Y + I+DD+ R WV F++ K+ F + ++++
Sbjct: 97 LDYIHSDLWGPSKVTSYGGRRYMMTIIDDFPRKVWVYFLRYKNETFPTFKKWRILVETQT 276
Query: 920 ELKILKVRSDHGGEFENEPFELFCEKHGILHEFSSPRTPQQNGVVERKNRTLQEMARTMI 979
+ K+ +D+ EF + F FC HGI + PR PQQNGV ER RTL E AR M+
Sbjct: 277 GKNVKKLITDN*LEFCSSDFNEFCTNHGIARHKTIPRNPQQNGVAERMIRTLLERARCML 456
Query: 980 HENNL--AKHFWAEAVNTSCYIQNRIYIRPMLEKTAYELFKGRRPNISYFHQFGCTCYIL 1037
L + W EA +T+C++ NR + K +++ G + S FGC Y L
Sbjct: 457 SNAGL*N*RDLWVEAASTACHLVNRSPHSALDFKVPEDIWSGNLVDYSNLRIFGCPAYAL 636
Query: 1038 NTKDYLKKFDAKAQRGIFLGYSERSKAYRVYNSE 1071
K +A IFL Y+ SK YR++ S+
Sbjct: 637 VNDG---KLAPRAGECIFLSYASESKGYRLWCSD 729
>BG587170 similar to PIR|F86470|F8 probable retroelement polyprotein [imported]
- Arabidopsis thaliana, partial (13%)
Length = 718
Score = 117 bits (293), Expect = 4e-26
Identities = 82/247 (33%), Positives = 129/247 (52%)
Frame = -3
Query: 757 VNKDDKSITFKGKRVENVYKINFSDLADQKVVCLLSMNDKKWVWHKRLGHANWRLISKIS 816
V K D + K V N YK +F+ + S+N K +WH RLGH + R ++ +
Sbjct: 680 VTKGDLYMLEKLDPVSN-YKCSFTSSS--------SLN-KDALWHARLGHPHGRALNLM- 534
Query: 817 KLQLVKGLPNIDYHSDALCGACQKGKIVKSSFKSKDIVSTSRPLELLHIDLFGPVNTASL 876
LP + + + C AC GK K+ F V + +L++ DL+ + S
Sbjct: 533 -------LPGVVFENKN-CEACILGKHCKNVFPRTSTVYENC-FDLIYTDLW-TAPSLSR 384
Query: 877 YGSKYGLVIVDDYSRWTWVKFIKSKDYACEVFSSFCTQIQSEKELKILKVRSDHGGEFEN 936
KY + +D+ S++TW+ I SKD + F +F + + KI +RSD+GGE+ +
Sbjct: 383 DNHKYFVTFIDEKSKYTWLTLIPSKDRVIDAFKNFQAYVTNHYHAKIKILRSDNGGEYTS 204
Query: 937 EPFELFCEKHGILHEFSSPRTPQQNGVVERKNRTLQEMARTMIHENNLAKHFWAEAVNTS 996
F+ + HGILH+ S P TPQQNGV +RKN+ L E+AR+++ + N V+T+
Sbjct: 203 YAFKSHLDHHGILHQTSCPYTPQQNGVAKRKNKHLMEVARSLMFQAN---------VSTA 51
Query: 997 CYIQNRI 1003
CY+ N I
Sbjct: 50 CYLINWI 30
>BI262917 weakly similar to GP|19920130|g Putative retroelement {Oryza sativa}
[Oryza sativa (japonica cultivar-group)], partial (8%)
Length = 426
Score = 102 bits (254), Expect(2) = 2e-22
Identities = 50/113 (44%), Positives = 69/113 (60%)
Frame = +1
Query: 957 TPQQNGVVERKNRTLQEMARTMIHENNLAKHFWAEAVNTSCYIQNRIYIRPMLEKTAYEL 1016
TPQQNGV ER NRTL E R M+ +AK FWAEAV T+CY+ NR + KT E+
Sbjct: 85 TPQQNGVAERMNRTLLERTRAMLKTAGMAKSFWAEAVKTACYVINRSPSTVIDLKTPMEM 264
Query: 1017 FKGRRPNISYFHQFGCTCYILNTKDYLKKFDAKAQRGIFLGYSERSKAYRVYN 1069
+KG+ + S H FGC Y++ K D K+++ IFLGY++ K Y +++
Sbjct: 265 WKGKPVDYSSLHVFGCPVYVMYNSQERTKLDPKSRKCIFLGYADNVKGYXLWD 423
Score = 23.5 bits (49), Expect(2) = 2e-22
Identities = 8/22 (36%), Positives = 15/22 (67%)
Frame = +3
Query: 932 GEFENEPFELFCEKHGILHEFS 953
GE+ + F FC++ GI+ +F+
Sbjct: 9 GEYVDGEFLAFCKQEGIVRQFT 74
>BG586293 weakly similar to PIR|E84473|E84 probable retroelement pol
polyprotein [imported] - Arabidopsis thaliana, partial
(7%)
Length = 763
Score = 84.3 bits (207), Expect = 3e-16
Identities = 42/86 (48%), Positives = 59/86 (67%)
Frame = +2
Query: 1239 LVPKPFQKNIIETKWVFRNKLNEQGEVTRNKARLVAQGYSQQEGINYTETFAPVARLETI 1298
LV KP I +W+++ K NE G + + KARLVA+GY +Q+GI++ E FAPV R+ETI
Sbjct: 65 LVKKPTGVKPIGLRWIYKIKRNEDGTLIKYKARLVAKGYVKQQGIDFDEVFAPVVRIETI 244
Query: 1299 RLLLSYAINHGIILYQMDVKSVFLNG 1324
LLL+ A +G ++ +DVK FLNG
Sbjct: 245 *LLLALAATNGC*IHHIDVKIAFLNG 322
>BE123913 weakly similar to GP|22093573|d polyprotein {Oryza sativa (japonica
cultivar-group)}, partial (8%)
Length = 503
Score = 81.3 bits (199), Expect = 3e-15
Identities = 45/125 (36%), Positives = 71/125 (56%), Gaps = 3/125 (2%)
Frame = +1
Query: 1359 QAPRAWYDRLSNFLIKNDFERGQVDTTLFRR---TLKKDILIVQIYVDDIIFGSTNASLC 1415
Q+PR W+DR + + K + + Q D +F + T+KK ILIV YVDDI +
Sbjct: 1 QSPRDWFDRFT*VVKKFGYIQCQTDHAMFIKHSSTVKKAILIV--YVDDIFLTGDHGK*I 174
Query: 1416 KEFSKLMQDEFEMSMMGELKFFLGIQINQSKEGVYVHQTKYTKELLKKFKLEDCKVMNTP 1475
K L+ +EFE+ +G LK+FLG+++ + K+G + Q KY +LLK+ ++ CK + P
Sbjct: 175 KRLKNLLAEEFEIKDLGNLKYFLGMEVARWKKGSSISQRKYVLDLLKETRMIGCKTIRDP 354
Query: 1476 MHPTC 1480
C
Sbjct: 355 YGCNC 369
Score = 53.5 bits (127), Expect = 6e-07
Identities = 25/56 (44%), Positives = 35/56 (61%)
Frame = +2
Query: 1468 DCKVMNTPMHPTCTLSKEDTGTVVDQKLYRGMIGSLLYLTASRPDILFSVCLCARF 1523
D K TPM T L D GT+VD+ Y+ ++G L+YL+ +RPDI F VC ++F
Sbjct: 332 DVKPSETPMDATVKLGTLDNGTLVDKGRYQRLVGKLIYLSHTRPDISFVVCTMSQF 499
>BG586273 weakly similar to PIR|F86470|F8 probable retroelement polyprotein
[imported] - Arabidopsis thaliana, partial (7%)
Length = 705
Score = 80.5 bits (197), Expect = 5e-15
Identities = 55/210 (26%), Positives = 98/210 (46%), Gaps = 8/210 (3%)
Frame = -2
Query: 996 SCYIQNRIYIRPMLEKTAYELFKGRRPNISYFHQFGCTCYILNTKDYLKKFDAKAQRGIF 1055
+CY+ NRI R + ++ +E+ R+P+++Y FGC CY+L + K +A++++ +F
Sbjct: 704 ACYLINRIPTRVLKDQAPFEVLNQRKPSLTYMRVFGCLCYVLVPGELRNKLEARSRKAMF 525
Query: 1056 LGYSERSKAYRVYNSETQCVEESMHVKFDDREPGSKTSEQSESNAGTTDSEDASESDQPS 1115
+GYS K Y+ Y+ E + V S VKF + + Q + T+D
Sbjct: 524 IGYSTTQKGYKCYDPEARRVLVSRDVKFIEERGYYEEKNQEDLRDLTSDKAGVLRVILEG 345
Query: 1116 DSEKYTKVESSPEAEITPEAESNS--EAESSPIVQNESASE-DFQDNTQQVI-----QPK 1167
K + +S+ + PE SN A +P + +E SE + Q+N Q+ + +
Sbjct: 344 LGIKMNQDQSTRSRQ--PEESSNEPRRAAQTPHLDHEGGSEPETQENGQEGVGLSEEGEE 171
Query: 1168 FKHKSSHPEELIIGSKDSPRRTRSHFRQEE 1197
F S H E G ++ + Q+E
Sbjct: 170 FNSGSHHQGEQDQGDEEHHSEAQEPHIQDE 81
>BF650113 weakly similar to GP|4753889|emb| Tpv2-1c {Phaseolus vulgaris},
partial (13%)
Length = 494
Score = 67.8 bits (164), Expect = 3e-11
Identities = 36/74 (48%), Positives = 49/74 (65%), Gaps = 3/74 (4%)
Frame = +1
Query: 1510 RPDILFSVCLCARFQSDPRESHLTAVKRIFRYLKGTTNLGLLY---RKSLDYKLIGFCDA 1566
RPDI +SV + ++F DPR+ HL A RI RY++GT GLL+ KS Y+LI + D+
Sbjct: 121 RPDICYSVSVISKFMHDPRKPHLIAANRILRYVRGTMEYGLLFPYGAKSEVYELICYSDS 300
Query: 1567 DYAGDRIERKSTSG 1580
D+ GD R+STSG
Sbjct: 301 DWCGD---RRSTSG 333
>CB893680 weakly similar to GP|1167523|db ORF(AA 1-1338) {Nicotiana tabacum},
partial (7%)
Length = 780
Score = 62.4 bits (150), Expect = 1e-09
Identities = 38/96 (39%), Positives = 52/96 (53%)
Frame = -2
Query: 1289 FAPVARLETIRLLLSYAINHGIILYQMDVKSVFLNGVIEEEVYVKQPPGFEDLKHPDHVY 1348
F P+ +L TI LLS + L +DVK+ FL G + E++Y+ QP GF + V
Sbjct: 554 FVPIVKLNTIMFLLSIVAIENLYLE*LDVKTAFLRGDLVEDIYMHQPEGF-S*EVGKMVG 378
Query: 1349 KLKKSLYGLKQAPRAWYDRLSNFLIKNDFERGQVDT 1384
KLKKS+YGLKQ PR L + F R ++ T
Sbjct: 377 KLKKSMYGLKQGPRQCI*SLKALCTRKGF-RSEIST 273
>BG644677 weakly similar to GP|7682800|gb| Hypothetical protein T15F17.l
{Arabidopsis thaliana}, partial (3%)
Length = 539
Score = 58.2 bits (139), Expect = 3e-08
Identities = 32/88 (36%), Positives = 47/88 (53%)
Frame = -3
Query: 1504 LYLTASRPDILFSVCLCARFQSDPRESHLTAVKRIFRYLKGTTNLGLLYRKSLDYKLIGF 1563
+ LT P+I FS+ L +R+ S P H +K I +YLKG ++GL Y K LIG+
Sbjct: 531 ILLTLQGPNITFSINLLSRYSSAPTMRH*NGIKHICKYLKGIIDMGLFYSKDCSPDLIGY 352
Query: 1564 CDADYAGDRIERKSTSGNCQFLGENLIS 1591
+A Y D + +S +G G +IS
Sbjct: 351 VNA*YLSDPHKARS*TGYIFTCGNTVIS 268
>BG587174 similar to PIR|A47759|A4775 retrovirus-related reverse transcriptase
homolog - rape retrotransposon copia-like (fragment),
partial (84%)
Length = 249
Score = 57.8 bits (138), Expect = 3e-08
Identities = 25/46 (54%), Positives = 33/46 (71%)
Frame = -1
Query: 1326 IEEEVYVKQPPGFEDLKHPDHVYKLKKSLYGLKQAPRAWYDRLSNF 1371
+EE++Y+ QP GF DHV KL+KSLYGLKQ+PR WY R ++
Sbjct: 246 LEEKIYMTQPEGFLFPGKEDHVCKLRKSLYGLKQSPRQWYKRFDSY 109
>TC92908 similar to GP|10140704|gb|AAG13538.1 putative gag-pol polyprotein
{Oryza sativa}, partial (1%)
Length = 638
Score = 55.5 bits (132), Expect = 2e-07
Identities = 55/203 (27%), Positives = 92/203 (45%), Gaps = 7/203 (3%)
Frame = +2
Query: 1122 KVESSPEAEITPEAESNSEAESSPIVQNESASEDFQDNTQQVIQPKFKHKSSHPEELIIG 1181
+VE+S EI + +S+ E + E + FQ+ TQ K K SH L+
Sbjct: 26 QVEASDLEEIQEDFKSHGEVQEESNYIEEI--KGFQEPTQL---RKIKE*ESH---LVTL 181
Query: 1182 SKDSPRRTRSHF-------RQEESLIGLLSIIEPKTVEEALSDDGWILAMQEELNQFQRN 1234
+ S H+ + SL + S IEPK +A W+ AM++E+ + N
Sbjct: 182 NSSSKSYHIFHYVGYSFSAKHRASLAAITSNIEPKNYVQAAQ*QEWLAAMEQEIQVLEEN 361
Query: 1235 DVWDLVPKPFQKNIIETKWVFRNKLNEQGEVTRNKARLVAQGYSQQEGINYTETFAPVAR 1294
+ L P K ++ + V++ GE+ + KA+LVA+ + Q EG ++ + +
Sbjct: 362 NTSTLEPLREGKKWVDCRPVYKIIHKANGEIEKYKAQLVAKDFVQVEGEDF*D-LCLSNK 538
Query: 1295 LETIRLLLSYAINHGIILYQMDV 1317
+ R LL+ A G L+ MDV
Sbjct: 539 DDNCRCLLTIAAAKG*QLHLMDV 607
>AJ499215 weakly similar to GP|6642775|gb| gag-pol polyprotein {Vitis
vinifera}, partial (18%)
Length = 567
Score = 53.5 bits (127), Expect = 6e-07
Identities = 38/134 (28%), Positives = 59/134 (43%), Gaps = 10/134 (7%)
Frame = +3
Query: 641 GVKPKVLNDQKPLSIHPKVCLRA------REKQRSWYLDSGCSRHMTGEKALFLTLTMKD 694
G P N KP K L A ++ + W +DSGC+ HMT ++ LF L
Sbjct: 3 GDSPSHKNKHKPHQ*GSKEQLLAMSTFATKQPSKYWLIDSGCTHHMTHDRDLFKELNKST 182
Query: 695 GGEVKFGGNQTGKIIGTGTIGNSSIS----INNVWLVDGLKHNLLSISQFCDNEYDVTFS 750
+V+ ++ G GT+ S S I+NV L +LLS+ Q Y V F
Sbjct: 183 ISKVRMLNGAHIEVEGIGTVLVKSHSGYKQISNVLYAPKLNQSLLSVPQLLTKGYKVLFE 362
Query: 751 KTNCTLVNKDDKSI 764
C + ++++K +
Sbjct: 363 HEKCVIKDQNNKEV 404
>TC76514 homologue to PIR|T09648|T09648 nucleolin homolog nuM1 - alfalfa,
partial (61%)
Length = 1288
Score = 50.1 bits (118), Expect = 7e-06
Identities = 58/238 (24%), Positives = 94/238 (39%), Gaps = 6/238 (2%)
Frame = +3
Query: 174 LQILKKSYVASDHVSKILRSLPSRWRPKVTAIEEAKDLNTLSVE-DLVSSLKVHEMSLNE 232
L+++KK +S+ S+ P P + AK N + + S + S ++
Sbjct: 267 LKVVKKEESSSEESSESEDEQPVVKAPAPSKKTPAKKGNVKKAQPETTSEESDSDSSSSD 446
Query: 233 HETSKKSKSIALPSKGKTSKSSKAYKASESEEESPDGDSDEDQS-VKMAMLSNKLEYLAR 291
E KK S A+PSK + S+ A K SEEE + SDED+ A+ S A+
Sbjct: 447 EEEVKKPVSKAVPSK---NGSAPAKKVDTSEEEDSEESSDEDKKPAAKAVPSKNGSAPAK 617
Query: 292 KQKKFLSKRGSYKNSKKEDQKGCFNC--KKPGHFIADCPDLQK--EKFKGKSKKSSFNSS 347
K + +ED+K K G A D + E + ++ ++
Sbjct: 618 KAASDEEDTDESSDEDEEDEKPAAKAVPSKNGSVPAKKADTESSDEDSESSDEEDKKPAA 797
Query: 348 KFRKQIKKSLMATWEDLDSESGSDKEEADDDAKAAMGLVATVSSEAVSEAESDSEDEN 405
K K + D ES + +E D+DAK A ++SD ED++
Sbjct: 798 KASKNVSAPTKKAASSSDEESDEESDE-DEDAKPVSKPAAVAKKSKKDSSDSDDEDDD 968
>CB894419 weakly similar to GP|10177935|db copia-type polyprotein {Arabidopsis
thaliana}, partial (2%)
Length = 170
Score = 50.1 bits (118), Expect = 7e-06
Identities = 24/55 (43%), Positives = 35/55 (63%)
Frame = +3
Query: 1455 KYTKELLKKFKLEDCKVMNTPMHPTCTLSKEDTGTVVDQKLYRGMIGSLLYLTAS 1509
K ++LKKFK+E K ++TP+ L++E G VD Y+ +IGSL YLTA+
Sbjct: 6 KCASDILKKFKMEHSKPISTPVEEKLKLTRESDGKRVDSTHYKSLIGSLRYLTAT 170
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.314 0.131 0.371
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 44,763,059
Number of Sequences: 36976
Number of extensions: 599434
Number of successful extensions: 2888
Number of sequences better than 10.0: 98
Number of HSP's better than 10.0 without gapping: 2771
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 2866
length of query: 1591
length of database: 9,014,727
effective HSP length: 109
effective length of query: 1482
effective length of database: 4,984,343
effective search space: 7386796326
effective search space used: 7386796326
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.9 bits)
S2: 65 (29.6 bits)
Medicago: description of AC139344.2


