

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC139344.1 + phase: 0
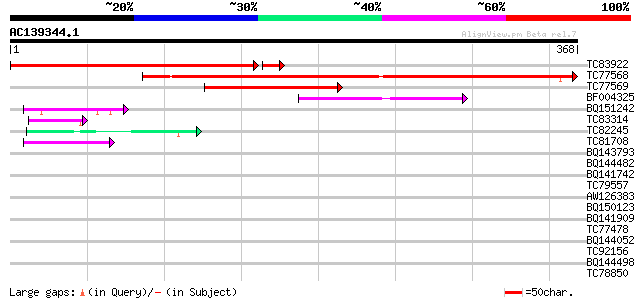
(368 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC83922 similar to GP|15809990|gb|AAL06922.1 AT5g60530/muf9_180 ... 352 e-101
TC77568 weakly similar to GP|9857516|gb|AAG00871.1| Unknown prot... 294 4e-80
TC77569 weakly similar to GP|9757754|dbj|BAB08235.1 contains sim... 123 1e-28
BF004325 weakly similar to GP|11994465|dbj contains similarity t... 79 3e-15
BQ151242 similar to PIR|G86292|G86 hypothetical protein AAF82153... 44 8e-05
TC83314 similar to GP|10177135|dbj|BAB10425. gene_id:K2A18.28~pi... 43 2e-04
TC82245 similar to GP|16648720|gb|AAL25552.1 AT4g21620/F17L22_80... 42 5e-04
TC81708 similar to GP|12083536|gb|AAG48845.1 hypothetical protei... 41 6e-04
BQ143793 weakly similar to GP|6523547|emb| hydroxyproline-rich g... 40 0.001
BQ144482 similar to GP|6523547|emb| hydroxyproline-rich glycopro... 40 0.002
BQ141742 weakly similar to PIR|T10798|T107 pherophorin-S - Volvo... 39 0.003
TC79557 similar to GP|12039289|gb|AAG46079.1 hypothetical protei... 39 0.004
AW126383 weakly similar to SP|P34480|NAGA_ Putative N-acetylgluc... 38 0.005
BQ150123 similar to GP|21109992|gb| TonB-dependent receptor {Xan... 38 0.005
BQ141909 weakly similar to GP|14253159|emb VMP3 protein {Volvox ... 38 0.007
TC77478 similar to GP|9884651|dbj|BAB11979.1 MGDG synthase type ... 37 0.009
BQ144052 weakly similar to GP|21327989|dbj contains ESTs AU09387... 37 0.012
TC92156 similar to GP|15724296|gb|AAL06541.1 AT5g61660/k11j9_180... 36 0.021
BQ144498 similar to GP|6330472|dbj KIAA1205 protein {Homo sapien... 35 0.060
TC78850 homologue to GP|10177413|dbj|BAB10544. fibrillarin-like ... 35 0.060
>TC83922 similar to GP|15809990|gb|AAL06922.1 AT5g60530/muf9_180
{Arabidopsis thaliana}, partial (21%)
Length = 619
Score = 352 bits (903), Expect(2) = e-101
Identities = 161/161 (100%), Positives = 161/161 (100%)
Frame = +1
Query: 1 MEAAIAQGQGNGNGNGNNGNGKGNGNGNGNNGNGKGNDKEEGKEKGKEKEKAPKKKKAPK 60
MEAAIAQGQGNGNGNGNNGNGKGNGNGNGNNGNGKGNDKEEGKEKGKEKEKAPKKKKAPK
Sbjct: 82 MEAAIAQGQGNGNGNGNNGNGKGNGNGNGNNGNGKGNDKEEGKEKGKEKEKAPKKKKAPK 261
Query: 61 EESSNYQQLQTLPSGQERAFCKANNTCHFATLVCPAECKTRKPKKNKKDKGCFIDCSSKC 120
EESSNYQQLQTLPSGQERAFCKANNTCHFATLVCPAECKTRKPKKNKKDKGCFIDCSSKC
Sbjct: 262 EESSNYQQLQTLPSGQERAFCKANNTCHFATLVCPAECKTRKPKKNKKDKGCFIDCSSKC 441
Query: 121 EATCKFRRANCDGYGSLCYDPRFVGGDGVMFYFHGAKGGNF 161
EATCKFRRANCDGYGSLCYDPRFVGGDGVMFYFHGAKGGNF
Sbjct: 442 EATCKFRRANCDGYGSLCYDPRFVGGDGVMFYFHGAKGGNF 564
Score = 33.5 bits (75), Expect(2) = e-101
Identities = 14/14 (100%), Positives = 14/14 (100%)
Frame = +3
Query: 165 SDDEFQINAHFIGT 178
SDDEFQINAHFIGT
Sbjct: 576 SDDEFQINAHFIGT 617
>TC77568 weakly similar to GP|9857516|gb|AAG00871.1| Unknown protein
{Arabidopsis thaliana}, partial (63%)
Length = 1206
Score = 294 bits (752), Expect = 4e-80
Identities = 135/285 (47%), Positives = 193/285 (67%), Gaps = 3/285 (1%)
Frame = +1
Query: 87 CHFATLVCPAECKTRKPKKNKKDKGCFIDCSSK-CEATCKFRRANCDGYGSLCYDPRFVG 145
C L CPA+C +R P N ++K C++DC S C+ CK R+ANC+G GS C DPRFVG
Sbjct: 64 CFLKKLPCPAQCPSRSPA-NPREKVCYLDCDSPMCKTQCKSRKANCNGRGSACLDPRFVG 240
Query: 146 GDGVMFYFHGAKGGNFAIVSDDEFQINAHFIGTRPQGRTRDYTWVQALSLMFDTHTLVIA 205
DG++FYFHG + +F++VSD QINA FIG RPQGR RDYTW+QAL ++FD+H I
Sbjct: 241 ADGIVFYFHGRRNEHFSLVSDTNLQINARFIGLRPQGRPRDYTWIQALGVLFDSHNFSIE 420
Query: 206 ANRVTQWNDNVDSLTVKWDGESVIIPTDDDAEWKTNGDEREVVVERTDDANSVRVTVSGL 265
A + W+D VD L + +DG+ ++IP + W++ +E ++ VERT + N V VT+ +
Sbjct: 421 ATPSSIWDDEVDHLKLSYDGKDLVIPEGHLSTWQS--EENQLRVERTSNENGVMVTIPEV 594
Query: 266 LEMDIRVRPIGEKENKAHNYQLPSDDAFAHLETQFKFKNPTDSIEGVLGQTYRPSYVSPV 325
E+ + V P+ +++++ HNYQ+P DD FAHL+ QFKF + +EGVLG+TY+P + +P
Sbjct: 595 AEISVNVVPVTKEDSRIHNYQIPDDDCFAHLDVQFKFYGLSSKVEGVLGRTYQPDFQNPA 774
Query: 326 KRGVAMPMMGGEDKYQTPSLFSTTCKLCRFQ--RPSTSQGLIAQY 368
K GVAMP++GGEDKY+T SL S C C F + S +G+ +Y
Sbjct: 775 KPGVAMPVVGGEDKYRTKSLVSADCLACIFSSAKDSVQEGMEIEY 909
>TC77569 weakly similar to GP|9757754|dbj|BAB08235.1 contains similarity to
root cap protein~gene_id:MUF9.15 {Arabidopsis thaliana},
partial (26%)
Length = 835
Score = 123 bits (309), Expect = 1e-28
Identities = 53/90 (58%), Positives = 67/90 (73%)
Frame = +3
Query: 127 RRANCDGYGSLCYDPRFVGGDGVMFYFHGAKGGNFAIVSDDEFQINAHFIGTRPQGRTRD 186
R+ANC+G GS C DPRFVG DG++FYFHG + +F++VSD QINA FIG RPQGR RD
Sbjct: 564 RKANCNGRGSACLDPRFVGADGIVFYFHGRRNEHFSLVSDTNLQINARFIGLRPQGRPRD 743
Query: 187 YTWVQALSLMFDTHTLVIAANRVTQWNDNV 216
YTW+QAL ++FD+H I A + W+D V
Sbjct: 744 YTWIQALGVLFDSHNFSIEATPSSIWDDEV 833
Score = 35.4 bits (80), Expect = 0.035
Identities = 16/40 (40%), Positives = 23/40 (57%), Gaps = 1/40 (2%)
Frame = +3
Query: 87 CHFATLVCPAECKTRKPKKNKKDKGCFIDCSS-KCEATCK 125
C L CPA+C +R P N ++K ++DC S C+ CK
Sbjct: 120 CFLKKLPCPAQCPSRSP-ANPREKVRYLDCDSPMCKTQCK 236
>BF004325 weakly similar to GP|11994465|dbj contains similarity to late
embryogenesis abundant protein~gene_id:MLD14.16
{Arabidopsis thaliana}, partial (8%)
Length = 357
Score = 79.0 bits (193), Expect = 3e-15
Identities = 35/110 (31%), Positives = 62/110 (55%)
Frame = +1
Query: 188 TWVQALSLMFDTHTLVIAANRVTQWNDNVDSLTVKWDGESVIIPTDDDAEWKTNGDEREV 247
TWVQ++ ++ D H L + A + W+D+VD L + +DGE + + + A+W+++G V
Sbjct: 1 TWVQSIVILLDNHQLFLGAQKTATWDDSVDRLAISFDGEPITLHESEGAKWESSG----V 168
Query: 248 VVERTDDANSVRVTVSGLLEMDIRVRPIGEKENKAHNYQLPSDDAFAHLE 297
R N++ V G ++ + PI E+ ++ HNY + DD AHL+
Sbjct: 169 SFGRETSTNNIIAEVEGNCKITAKGVPITEEHSRVHNYGITKDDCCAHLD 318
>BQ151242 similar to PIR|G86292|G86 hypothetical protein AAF82153.1
[imported] - Arabidopsis thaliana, partial (5%)
Length = 1371
Score = 44.3 bits (103), Expect = 8e-05
Identities = 30/81 (37%), Positives = 36/81 (44%), Gaps = 13/81 (16%)
Frame = +2
Query: 10 GNGNGNGNNG-----NGKGNGNGNGNNGNGKGNDKEEGKEKGKEKEKAPKK------KKA 58
GN G G+ G GKG G G G KG KEEG E E+ K K+ K+
Sbjct: 113 GNKKGRGSEGAGGRERGKGGGGGGERGGGQKGRRKEEGAEGAAERPKERKREGGGEAKQG 292
Query: 59 PKEESS--NYQQLQTLPSGQE 77
P E +S N Q + P G E
Sbjct: 293 PGERASTPNQPQKKPQPPGSE 355
>TC83314 similar to GP|10177135|dbj|BAB10425.
gene_id:K2A18.28~pir||T10240~strong similarity to
unknown protein {Arabidopsis thaliana}, partial (26%)
Length = 956
Score = 42.7 bits (99), Expect = 2e-04
Identities = 21/46 (45%), Positives = 25/46 (53%), Gaps = 8/46 (17%)
Frame = +1
Query: 13 NGNGNNGNGKGNGNGNGNNGNGKGNDKEEGKE--------KGKEKE 50
N N NGNG GNGNGNGN+G +GN + KG+E E
Sbjct: 817 NSNTPNGNGLGNGNGNGNDGGKQGNHNNHQRNYSHSGINMKGRESE 954
Score = 30.4 bits (67), Expect = 1.1
Identities = 15/35 (42%), Positives = 17/35 (47%)
Frame = +1
Query: 14 GNGNNGNGKGNGNGNGNNGNGKGNDKEEGKEKGKE 48
GN N N N N NGNG GN G + GK+
Sbjct: 781 GNEANQNQNILANSNTPNGNGLGNGNGNGNDGGKQ 885
Score = 27.7 bits (60), Expect = 7.4
Identities = 17/43 (39%), Positives = 21/43 (48%), Gaps = 2/43 (4%)
Frame = +1
Query: 4 AIAQGQGNGNGNGNNGNGKGNGNG-NGNN-GNGKGNDKEEGKE 44
A ++ GN N N N N NGN GNG GN + GK+
Sbjct: 757 AASKQPNEGNEANQNQNILANSNTPNGNGLGNGNGNGNDGGKQ 885
>TC82245 similar to GP|16648720|gb|AAL25552.1 AT4g21620/F17L22_80
{Arabidopsis thaliana}, partial (67%)
Length = 604
Score = 41.6 bits (96), Expect = 5e-04
Identities = 35/120 (29%), Positives = 40/120 (33%), Gaps = 7/120 (5%)
Frame = +3
Query: 12 GNGNGNNGNGKGNGNGNG-NNGNGKGNDKEEGKEKGKEKEKAPKKKKAPKEESSNYQQLQ 70
G G G N G GNG GNG G G G G G K +
Sbjct: 267 GPGGGFNIPGFGNGFGNGVGGGYGSGYGGPNG---GHSKNGIVRPT-------------- 395
Query: 71 TLPSGQERAFCKANNTCHFATLVCPAECKTRKPKKNKK------DKGCFIDCSSKCEATC 124
CK C+ L CPA+C + + K GC IDC KC A C
Sbjct: 396 --------VVCKDKGPCYQKKLTCPAKCFSSFSRSGKGYGGGGGGGGCTIDCKKKCSAYC 551
>TC81708 similar to GP|12083536|gb|AAG48845.1 hypothetical protein {Oryza
sativa}, partial (6%)
Length = 605
Score = 41.2 bits (95), Expect = 6e-04
Identities = 21/60 (35%), Positives = 27/60 (45%), Gaps = 1/60 (1%)
Frame = +1
Query: 10 GNGNGNGNNGNGKGN-GNGNGNNGNGKGNDKEEGKEKGKEKEKAPKKKKAPKEESSNYQQ 68
GNGNGNG KGN NG+G+N NG G + G A + P + + Q
Sbjct: 424 GNGNGNGKGAPAKGNANNGHGSNNNGNGGHMGPMGQMGAMNHMAAMNRMGPMGQMGSMDQ 603
Score = 35.8 bits (81), Expect = 0.027
Identities = 16/33 (48%), Positives = 19/33 (57%), Gaps = 3/33 (9%)
Frame = +1
Query: 13 NGNGNNGNGKGNG---NGNGNNGNGKGNDKEEG 42
+G G NGNG G G GN NNG+G N+ G
Sbjct: 412 SGKGGNGNGNGKGAPAKGNANNGHGSNNNGNGG 510
Score = 30.8 bits (68), Expect = 0.87
Identities = 18/61 (29%), Positives = 24/61 (38%), Gaps = 1/61 (1%)
Frame = +1
Query: 7 QGQGNGNGNGNNGNGKGNGNGNGNNGNGKG-NDKEEGKEKGKEKEKAPKKKKAPKEESSN 65
+G+ G NGNG G G+ KG K G + G EK + KK K
Sbjct: 37 KGKSGNYGEVKNGNGGGKKGGDDGGKKNKGEKQKGGGGDWGDEKNSSKKKNGKGKNGGGG 216
Query: 66 Y 66
+
Sbjct: 217F 219
Score = 28.9 bits (63), Expect = 3.3
Identities = 21/61 (34%), Positives = 26/61 (42%), Gaps = 19/61 (31%)
Frame = +1
Query: 13 NGNGNNGNG--------------KGNG-----NGNGNNGNGKGNDKEEGKEKGKEKEKAP 53
NG G NG G KG G N NNG G ++ +GK+ GK+ EK
Sbjct: 187 NGKGKNGGGGFLVKFLGLGKKSKKGGGAADTTNKKKNNGGGGNSNNSKGKD-GKKGEKLD 363
Query: 54 K 54
K
Sbjct: 364 K 366
>BQ143793 weakly similar to GP|6523547|emb| hydroxyproline-rich glycoprotein
DZ-HRGP {Volvox carteri f. nagariensis}, partial (19%)
Length = 1100
Score = 40.0 bits (92), Expect = 0.001
Identities = 22/48 (45%), Positives = 24/48 (49%)
Frame = -1
Query: 7 QGQGNGNGNGNNGNGKGNGNGNGNNGNGKGNDKEEGKEKGKEKEKAPK 54
+G G G GNG G G G G G G G G DK K K KEK+ K
Sbjct: 278 KGGGMGRGNG--GEGGGGGEGGGREWGGGGGDK*SLKRKEKEKDGGDK 141
Score = 39.3 bits (90), Expect = 0.002
Identities = 24/73 (32%), Positives = 32/73 (42%), Gaps = 22/73 (30%)
Frame = -3
Query: 7 QGQGNGNGNGNNGNG----------------------KGNGNGNGNNGNGKGNDKEEGKE 44
+G G G G G G+G + G NGN G G+G E KE
Sbjct: 654 RGGGGGRGGGGRGDGIVVREVRVKRRKMDGANEERWGRERGRVNGNRGKGRGGTSER-KE 478
Query: 45 KGKEKEKAPKKKK 57
+GKEKE+ K+K+
Sbjct: 477 EGKEKEERGKRKE 439
Score = 35.8 bits (81), Expect = 0.027
Identities = 17/50 (34%), Positives = 25/50 (50%)
Frame = -2
Query: 2 EAAIAQGQGNGNGNGNNGNGKGNGNGNGNNGNGKGNDKEEGKEKGKEKEK 51
E+ QGQG G G G+G G G G GK K+ G+ +G+ + +
Sbjct: 532 ESKWEQGQGEGGDKRKKGRGEGKG-GKGKTQRGKRGGKKGGRRRGRRERE 386
Score = 33.5 bits (75), Expect = 0.13
Identities = 22/58 (37%), Positives = 27/58 (45%), Gaps = 3/58 (5%)
Frame = -3
Query: 8 GQGNGNGNGNNGNGKGNGNGNGNNGNGKGNDKEEGKEK---GKEKEKAPKKKKAPKEE 62
G+ G NGN G G+G G KEEGKEK GK KE ++K + E
Sbjct: 546 GRERGRVNGNRGKGRG----------GTSERKEEGKEKEERGKRKEGKGGERKGEEGE 403
Score = 32.3 bits (72), Expect = 0.30
Identities = 17/43 (39%), Positives = 23/43 (52%)
Frame = -1
Query: 8 GQGNGNGNGNNGNGKGNGNGNGNNGNGKGNDKEEGKEKGKEKE 50
G G G G G G + GNG G+G KEE +++G+E E
Sbjct: 137 G*GKGRGGGEGGM---DVMGNGGGRRGEGRKKEEREKRGEEGE 18
Score = 31.6 bits (70), Expect = 0.51
Identities = 20/53 (37%), Positives = 23/53 (42%), Gaps = 13/53 (24%)
Frame = -1
Query: 12 GNGNGNN------GNGKGNGNGNGNNGNGKGNDKEEG-------KEKGKEKEK 51
G G G + G G G GNG G G+G +E G K KEKEK
Sbjct: 314 GRGKGKDEII*EKGGGMGRGNGGEGGGGGEGGGREWGGGGGDK*SLKRKEKEK 156
Score = 29.3 bits (64), Expect = 2.5
Identities = 18/44 (40%), Positives = 23/44 (51%), Gaps = 3/44 (6%)
Frame = -3
Query: 5 IAQGQGNGN---GNGNNGNGKGNGNGNGNNGNGKGNDKEEGKEK 45
I +G+G+G G G G +G G G G G G KE+GK K
Sbjct: 285 IGEGRGDGKRKWGGGGGGRRRG-GEGMGRGGGG*MIFKEKGKGK 157
Score = 28.1 bits (61), Expect = 5.7
Identities = 16/62 (25%), Positives = 26/62 (41%), Gaps = 1/62 (1%)
Frame = -2
Query: 8 GQGNGNGNGNNGNGKGNGNGNGNNGNGKGNDKEEGK-EKGKEKEKAPKKKKAPKEESSNY 66
G+ G G G+G G GKG +GK ++GK K +++ +E Y
Sbjct: 547 GKRKGESKWEQGQGEGGDKRKKGRGEGKGG---KGKTQRGKRGGKKGGRRRGRREREMGY 377
Query: 67 QQ 68
+
Sbjct: 376 MK 371
Score = 27.7 bits (60), Expect = 7.4
Identities = 20/71 (28%), Positives = 26/71 (36%), Gaps = 21/71 (29%)
Frame = -2
Query: 8 GQGNGNGNGNNGN---------------GKGNGNGNGNNG------NGKGNDKEEGKEKG 46
G GNG G G + G+G G G G G G G K K +G
Sbjct: 220 GGGNGEGGGGINDL*RERKRKKMGGIR*GRGKGGGEGKGGWMLWGMGGGGGGKGGKKRRG 41
Query: 47 KEKEKAPKKKK 57
K+ K + K+
Sbjct: 40 KKGGKKERGKR 8
>BQ144482 similar to GP|6523547|emb| hydroxyproline-rich glycoprotein DZ-HRGP
{Volvox carteri f. nagariensis}, partial (16%)
Length = 993
Score = 39.7 bits (91), Expect = 0.002
Identities = 17/44 (38%), Positives = 24/44 (53%), Gaps = 4/44 (9%)
Frame = -1
Query: 8 GQGNG----NGNGNNGNGKGNGNGNGNNGNGKGNDKEEGKEKGK 47
G+G G G G G G+G G G G G+G + EG+E+G+
Sbjct: 354 GRGGGARGAGGRGGEGRGQGEAEGGGGGGRGRGAKEGEGRERGR 223
Score = 32.7 bits (73), Expect = 0.23
Identities = 16/40 (40%), Positives = 20/40 (50%), Gaps = 1/40 (2%)
Frame = -1
Query: 7 QGQGNGNGNGNNGNGKGNGNGNG-NNGNGKGNDKEEGKEK 45
+GQG G G G G+G G G G G G+ + G EK
Sbjct: 306 RGQGEAEGGGGGGRGRGAKEGEGRERGRGGGDRRGGGGEK 187
Score = 31.2 bits (69), Expect = 0.67
Identities = 16/55 (29%), Positives = 23/55 (41%), Gaps = 2/55 (3%)
Frame = -2
Query: 7 QGQGNGNGNGNNGNGKGNGNGNGNNGNGKGNDKE--EGKEKGKEKEKAPKKKKAP 59
+G G G G G G+G G G G+G K G G+ P+++ P
Sbjct: 287 RGGGGGGGGGGRKRGRGGRGGGGAEIEGEGGGKRWARGDFGGRGPVAGPRRRARP 123
>BQ141742 weakly similar to PIR|T10798|T107 pherophorin-S - Volvox carteri,
partial (9%)
Length = 1337
Score = 38.9 bits (89), Expect = 0.003
Identities = 17/38 (44%), Positives = 21/38 (54%)
Frame = +2
Query: 10 GNGNGNGNNGNGKGNGNGNGNNGNGKGNDKEEGKEKGK 47
G G GNG G G+G G +G G G G EG E+G+
Sbjct: 17 GGGGGNGGWGGGEGGGGVSGGLGLGVGGQPREGAERGR 130
>TC79557 similar to GP|12039289|gb|AAG46079.1 hypothetical protein {Oryza
sativa}, partial (45%)
Length = 754
Score = 38.5 bits (88), Expect = 0.004
Identities = 18/50 (36%), Positives = 22/50 (44%), Gaps = 6/50 (12%)
Frame = +1
Query: 81 CKANNTCHFATLVCPAECKTRKPKKNKK------DKGCFIDCSSKCEATC 124
CK C+ + CPA C T + K GC IDC KC A+C
Sbjct: 361 CKDRGPCYQKKVTCPARCFTSFSRSGKGYGGGGGGGGCTIDCKKKCTASC 510
>AW126383 weakly similar to SP|P34480|NAGA_ Putative
N-acetylglucosamine-6-phosphate deacetylase (EC
3.5.1.25) (GlcNAc 6-P deacetylase)., partial (11%)
Length = 525
Score = 38.1 bits (87), Expect = 0.005
Identities = 17/20 (85%), Positives = 17/20 (85%)
Frame = +1
Query: 11 NGNGNGNNGNGKGNGNGNGN 30
NGN NGN GNG GNGNGNGN
Sbjct: 127 NGNMNGN-GNGNGNGNGNGN 183
Score = 35.8 bits (81), Expect = 0.027
Identities = 16/29 (55%), Positives = 19/29 (65%)
Frame = +1
Query: 5 IAQGQGNGNGNGNNGNGKGNGNGNGNNGN 33
I G NGNGNGN GNG GNGN ++ +
Sbjct: 121 IQNGNMNGNGNGN-GNGNGNGNATSSSSS 204
>BQ150123 similar to GP|21109992|gb| TonB-dependent receptor {Xanthomonas
axonopodis pv. citri str. 306}, partial (2%)
Length = 1131
Score = 38.1 bits (87), Expect = 0.005
Identities = 20/67 (29%), Positives = 32/67 (46%), Gaps = 4/67 (5%)
Frame = +2
Query: 9 QGNGNGNGNNGNGKGNGNGNGNNGNGKGNDKEEGKEKGKEK----EKAPKKKKAPKEESS 64
+G G G G K NG GKG +EG++ +E+ +KAP+++ A E+
Sbjct: 5 EGRGGGGGTEKTEKRNGREGRKRTEGKGGGGKEGQKNKRERKDTIQKAPRERGAGSGEAK 184
Query: 65 NYQQLQT 71
+Q T
Sbjct: 185RMEQKST 205
>BQ141909 weakly similar to GP|14253159|emb VMP3 protein {Volvox carteri f.
nagariensis}, partial (13%)
Length = 1223
Score = 37.7 bits (86), Expect = 0.007
Identities = 19/49 (38%), Positives = 25/49 (50%), Gaps = 11/49 (22%)
Frame = +3
Query: 14 GNGNNGNGKGNGNGNG-----------NNGNGKGNDKEEGKEKGKEKEK 51
G G+ GN KGN GNG N G KG + G+E+G E+E+
Sbjct: 1035 GGGSRGNQKGNAGGNGDERESR*QKRKNEGEEKGGARRGGEERGVEEER 1181
Score = 36.6 bits (83), Expect = 0.016
Identities = 17/48 (35%), Positives = 24/48 (49%), Gaps = 3/48 (6%)
Frame = +1
Query: 7 QGQGNGNGNGNNGNGKGNGNG---NGNNGNGKGNDKEEGKEKGKEKEK 51
+G+ G G G G+G G G G G G G+G + E + G+ K K
Sbjct: 175 KGEAGGGGGGGGGSGGGVGEGKREEGGGGEGRGEEGEGEERGGRNKRK 318
Score = 33.5 bits (75), Expect = 0.13
Identities = 19/58 (32%), Positives = 27/58 (45%), Gaps = 6/58 (10%)
Frame = +1
Query: 10 GNGNGNGNNGNGKGNGNGNGNNGN----GKGNDKEEGKEKGKE--KEKAPKKKKAPKE 61
G G G G G G+ G N G G+ +GK +GKE +E+A K + +E
Sbjct: 253 GGGEGRGEEGEGEERGGRNKRKGEA*ERGRNGGGRQGKNRGKEWDREEAGGKWRGKRE 426
Score = 32.7 bits (73), Expect = 0.23
Identities = 15/45 (33%), Positives = 22/45 (48%), Gaps = 3/45 (6%)
Frame = +2
Query: 8 GQGNGNGNGNNGNGKGNGNGNGNNGNGKGNDKEEG---KEKGKEK 49
G+G G G + G+G G G G +G K+ G K +GK +
Sbjct: 197 GEGGGGAEGGSERGRGRRGGGGRGGGRRGRGKKGGGGTKGRGKRE 331
Score = 32.0 bits (71), Expect = 0.39
Identities = 15/50 (30%), Positives = 21/50 (42%)
Frame = +2
Query: 2 EAAIAQGQGNGNGNGNNGNGKGNGNGNGNNGNGKGNDKEEGKEKGKEKEK 51
E +G+G G G G +G G G G+G + G +G K K
Sbjct: 218 EGGSERGRGRRGGGGRGGGRRGRGKKGGGGTKGRGKRERGGGTEGGGKGK 367
>TC77478 similar to GP|9884651|dbj|BAB11979.1 MGDG synthase type A {Glycine
max}, partial (85%)
Length = 2229
Score = 37.4 bits (85), Expect = 0.009
Identities = 22/52 (42%), Positives = 27/52 (51%)
Frame = +1
Query: 3 AAIAQGQGNGNGNGNNGNGKGNGNGNGNNGNGKGNDKEEGKEKGKEKEKAPK 54
A++ G G G+G G G G GN G NGNG G + E K E +KA K
Sbjct: 733 ASLRVGDG---GDGGGGGGVGEGNDVGGNGNGVGVEDEGLGLKSGEGKKAKK 879
>BQ144052 weakly similar to GP|21327989|dbj contains ESTs AU093876(E1018)
AU093877(E1018)~unknown protein, partial (11%)
Length = 536
Score = 37.0 bits (84), Expect = 0.012
Identities = 27/61 (44%), Positives = 33/61 (53%), Gaps = 8/61 (13%)
Frame = -1
Query: 3 AAIAQGQG-NGNGNGNNGNGKGNG----NGNGNNGNGKGNDKEEGKEK---GKEKEKAPK 54
A I+ GQG G G G GNGKGNG G GN G +GN EEG + KE ++ +
Sbjct: 371 AGISGGQGIAGRGGGAMGNGKGNGGWGRKGGGNWGGCEGN--EEGGNR*AHWKEGDRVGR 198
Query: 55 K 55
K
Sbjct: 197 K 195
>TC92156 similar to GP|15724296|gb|AAL06541.1 AT5g61660/k11j9_180
{Arabidopsis thaliana}, partial (67%)
Length = 650
Score = 36.2 bits (82), Expect = 0.021
Identities = 18/51 (35%), Positives = 29/51 (56%), Gaps = 8/51 (15%)
Frame = +2
Query: 8 GQGNGNG-----NGNNGNGKGNGNGN---GNNGNGKGNDKEEGKEKGKEKE 50
G G+G+G G +G G G+G+GN G +G G+ N++ + KGK +
Sbjct: 335 GSGSGSGYGYGSGGAHGGGYGSGSGNSGGGGHGGGRTNNRSPSESKGKTNQ 487
Score = 29.6 bits (65), Expect = 1.9
Identities = 18/71 (25%), Positives = 28/71 (39%), Gaps = 21/71 (29%)
Frame = +2
Query: 8 GQGNGNGNGNNGNGKGNGNG---------------------NGNNGNGKGNDKEEGKEKG 46
G G+G+G+ +G G+G G G G G+G GN G G
Sbjct: 260 GYGSGSGHSPSGFGRGYGYGFGTGSGSGSGSGYGYGSGGAHGGGYGSGSGNSGGGGHGGG 439
Query: 47 KEKEKAPKKKK 57
+ ++P + K
Sbjct: 440 RTNNRSPSESK 472
>BQ144498 similar to GP|6330472|dbj KIAA1205 protein {Homo sapiens}, partial
(3%)
Length = 1284
Score = 34.7 bits (78), Expect = 0.060
Identities = 15/55 (27%), Positives = 27/55 (48%)
Frame = +3
Query: 2 EAAIAQGQGNGNGNGNNGNGKGNGNGNGNNGNGKGNDKEEGKEKGKEKEKAPKKK 56
E A +G+ G+G G G G+G G+G++ G G+ G + P+++
Sbjct: 201 ETAAEEGERGGDGGGWGGRGRGRAGGDGDHTRGAAACTVPGRAGGLGRAGGPRRR 365
>TC78850 homologue to GP|10177413|dbj|BAB10544. fibrillarin-like
{Arabidopsis thaliana}, partial (87%)
Length = 1177
Score = 34.7 bits (78), Expect = 0.060
Identities = 18/46 (39%), Positives = 22/46 (47%), Gaps = 4/46 (8%)
Frame = +3
Query: 8 GQGNGNGNGNN----GNGKGNGNGNGNNGNGKGNDKEEGKEKGKEK 49
G+G G G G + G G+G G G G G G+G G KG K
Sbjct: 120 GRGFGGGRGGDFKPRGGGRGFGGGRGARGGGRGRGGGRGGMKGGSK 257
Score = 32.3 bits (72), Expect = 0.30
Identities = 15/46 (32%), Positives = 22/46 (47%)
Frame = +3
Query: 1 MEAAIAQGQGNGNGNGNNGNGKGNGNGNGNNGNGKGNDKEEGKEKG 46
+ A +G+G G G + G+G G G G G G+ K G +G
Sbjct: 42 LNMAPPRGRGGFGGRGGDRGGRGGGGGRGFGGGRGGDFKPRGGGRG 179
Score = 30.8 bits (68), Expect = 0.87
Identities = 18/49 (36%), Positives = 22/49 (44%), Gaps = 4/49 (8%)
Frame = +3
Query: 8 GQGNGNGNGNNGNGKGNGNGNGNNGNGKGNDKEEGKEKG----KEKEKA 52
G+G G G G G G+G G G G G E + +G K KE A
Sbjct: 171 GRGFGGGRGARGGGRGRGGGRGGMKGGSKVVVEPHRHEGIFIAKGKEDA 317
Score = 28.1 bits (61), Expect = 5.7
Identities = 12/39 (30%), Positives = 18/39 (45%)
Frame = +3
Query: 8 GQGNGNGNGNNGNGKGNGNGNGNNGNGKGNDKEEGKEKG 46
G+G G G G+G G G G + +G + G +G
Sbjct: 81 GRGGDRGGRGGGGGRGFGGGRGGDFKPRGGGRGFGGGRG 197
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.314 0.133 0.404
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 12,036,926
Number of Sequences: 36976
Number of extensions: 188209
Number of successful extensions: 1872
Number of sequences better than 10.0: 170
Number of HSP's better than 10.0 without gapping: 1235
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1656
length of query: 368
length of database: 9,014,727
effective HSP length: 97
effective length of query: 271
effective length of database: 5,428,055
effective search space: 1471002905
effective search space used: 1471002905
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.9 bits)
S2: 59 (27.3 bits)
Medicago: description of AC139344.1


