

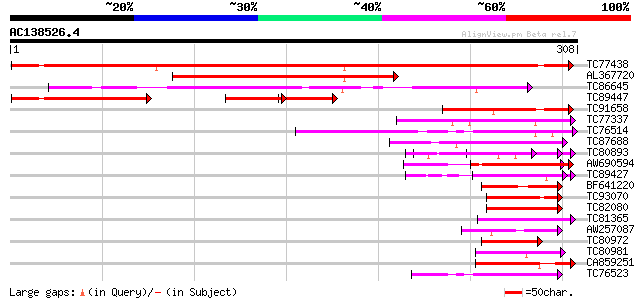
BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC138526.4 + phase: 0
(308 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC77438 homologue to PIR|T06807|T06807 nucleosome assembly prote... 384 e-107
AL367720 similar to PIR|S60892|S60 nucleosome assembly protein 1... 154 3e-38
TC86645 similar to PIR|H86321|H86321 hypothetical protein AAF271... 107 4e-24
TC89447 similar to PIR|S60892|S60892 nucleosome assembly protein... 91 7e-19
TC91658 similar to GP|14329812|emb|CAC40753. putative nucleosome... 75 4e-14
TC77337 weakly similar to GP|4019275|gb|AAC95573.1| orf 48 {Atel... 61 5e-10
TC76514 homologue to PIR|T09648|T09648 nucleolin homolog nuM1 - ... 51 6e-07
TC87688 similar to GP|10177535|dbj|BAB10930. gene_id:K1F13.21~un... 50 8e-07
TC80893 similar to GP|4557063|gb|AAD22502.1| expressed protein {... 47 9e-06
AW690594 similar to GP|23498163|emb hypothetical protein {Plasmo... 47 9e-06
TC89427 weakly similar to GP|4557063|gb|AAD22502.1| expressed pr... 46 2e-05
BF641220 weakly similar to PIR|G86203|G86 probable N-arginine di... 45 5e-05
TC93070 weakly similar to GP|17065230|gb|AAL32769.1 Unknown prot... 45 5e-05
TC82080 similar to GP|9757854|dbj|BAB08488.1 contains similarity... 44 8e-05
TC81365 similar to GP|14579399|gb|AAK69274.1 unknown {Glycine ma... 44 1e-04
AW257087 homologue to GP|6683126|dbj KIAA0339 protein {Homo sapi... 44 1e-04
TC80972 weakly similar to GP|23496972|gb|AAN36523.1 hypothetical... 43 1e-04
TC80981 similar to GP|8698730|gb|AAF78488.1| ESTs gb|Z18526 gb|... 43 2e-04
CA859251 similar to GP|21305823|gb DNA polymerase I {Hz-1 insect... 43 2e-04
TC76523 homologue to PIR|T09648|T09648 nucleolin homolog nuM1 - ... 43 2e-04
>TC77438 homologue to PIR|T06807|T06807 nucleosome assembly protein 1 -
garden pea, partial (93%)
Length = 1607
Score = 384 bits (987), Expect = e-107
Identities = 199/320 (62%), Positives = 248/320 (77%), Gaps = 15/320 (4%)
Frame = +2
Query: 2 IQALTQQRYKALQYLFASPIVEKRLEVLKEIQKEHDDLKAKFFEERAALEAKYQLLYQPL 61
IQ+L Q L+ L SP+V KR+EVL+EIQ EHD+L+AKF EERAALEAKYQ LYQPL
Sbjct: 221 IQSLAGQHSDVLESL--SPVVRKRVEVLREIQGEHDELEAKFLEERAALEAKYQKLYQPL 394
Query: 62 YTKRYEIVNGLAEVEGA------DIDEPEVKGVPYFWLVALQNNDEIADEITERDEDALK 115
YTKRY+IVNG+ EVEGA D +E KGVP FWL A++NND +++E+TERDE ALK
Sbjct: 395 YTKRYDIVNGVTEVEGAAMETTADAEEDNEKGVPSFWLNAMKNNDVLSEEVTERDEAALK 574
Query: 116 YLKDIKYTRTTEPQGFKLEFFFDSNPYFSNTILTKTYHMVDEDEFIMEKAIGTEIEWLPG 175
+LKDIK++R +P+GFKLEFFFD+NPYFSN+ILTKTYHMVDEDE I+EKAIGTEI+WLPG
Sbjct: 575 FLKDIKWSRIDDPKGFKLEFFFDTNPYFSNSILTKTYHMVDEDEPILEKAIGTEIQWLPG 754
Query: 176 KSLTE-------KKDPNTAKQTIETEKFDSFFNFFNSLEIPKDGMGVDEEADEELQNEME 228
K LT+ KK AK +TE +SFFNFFN E+P+D +DE+ EELQN+ME
Sbjct: 755 KCLTQKVLKKKPKKGAKNAKPITKTETCESFFNFFNPPEVPEDDEDIDEDMAEELQNQME 934
Query: 229 HDYDIGSTIRDKIIPNAVSWYTGEASDGES-GDLDDDDDDEDEE-DDEEEVEVEKEEEDD 286
DYDIGSTIRDKIIP+AVSW+TGEA+ GE GDLDD+DDDED++ +D++E E E E+EDD
Sbjct: 935 QDYDIGSTIRDKIIPHAVSWFTGEAAQGEEFGDLDDEDDDEDDDAEDDDEDEDEDEDEDD 1114
Query: 287 NYGLDDDEEDDNEDVNNPKK 306
+ +D+EE + ++ KK
Sbjct: 1115D---EDEEETKTKKKSSAKK 1165
>AL367720 similar to PIR|S60892|S60 nucleosome assembly protein 1 - soybean,
partial (36%)
Length = 402
Score = 154 bits (390), Expect = 3e-38
Identities = 74/130 (56%), Positives = 96/130 (72%), Gaps = 7/130 (5%)
Frame = +1
Query: 89 PYFWLVALQNNDEIADEITERDEDALKYLKDIKYTRTTEPQGFKLEFFFDSNPYFSNTIL 148
P FWL A++ N+ +EITERDE+ALKYLKDIK+++ P+G KLEF+FDSNPYF N++L
Sbjct: 7 PSFWLTAMKTNEI*GEEITERDEEALKYLKDIKWSKLDNPKGNKLEFYFDSNPYFQNSVL 186
Query: 149 TKTYHMVDEDEFIMEKAIGTEIEWLPGKSLTE-------KKDPNTAKQTIETEKFDSFFN 201
TKTYHM+++D+ I+EK+IGTEI+W PGK T+ KK AK I+TEK DSFFN
Sbjct: 187 TKTYHMIEDDDPILEKSIGTEIDWHPGKC*TQKVMTKKPKKGSQNAKPIIKTEKCDSFFN 366
Query: 202 FFNSLEIPKD 211
F IP+D
Sbjct: 367 IFKPPHIPED 396
>TC86645 similar to PIR|H86321|H86321 hypothetical protein AAF27100.1
[imported] - Arabidopsis thaliana, partial (76%)
Length = 1116
Score = 107 bits (268), Expect = 4e-24
Identities = 78/276 (28%), Positives = 131/276 (47%), Gaps = 13/276 (4%)
Frame = +3
Query: 22 VEKRLEVLKEIQKEHDDLKAKFFEERAALEAKYQLLYQPLYTKRYEIVNGLAEVEGADID 81
+EK E+ EI+K +++ K E +E KY + +P+Y KR +++
Sbjct: 144 IEKLQEIQDEIEKINEEASDKVLE----IEQKYNEVRKPVYDKRNDVI------------ 275
Query: 82 EPEVKGVPYFWLVALQNNDEIADEITERDEDALKYLKDIKYTRTTEPQ-GFKLEFFFDSN 140
K +P FWL A ++ + D + E D+ K+L ++ + + G+ + F F+SN
Sbjct: 276 ----KSIPDFWLTAFLSHPVLGDLLNEEDQKIFKHLISLEVEDHKDVKSGYSITFNFNSN 443
Query: 141 PYFSNTILTKTYHMVDEDEFIMEKAIGTEIEWLPGKSLT-----EKKDPNTAKQTIETEK 195
PYF ++ L KT+ ++E K T I+W GK + EKK A I
Sbjct: 444 PYFEDSKLVKTFTFLEEGT---TKLTATPIKWKEGKGIPNGVSHEKKGNKRAASDI---- 602
Query: 196 FDSFFNFFNSLEIPKDGMGVDEEADEELQNEMEHDYD-IGSTIRDKIIPNAVSWYTGE-- 252
SFF +F D E ++EM +D I I+D + PN ++++ E
Sbjct: 603 --SFFTWF---------------CDTEQKDEMGDIHDEIAEMIKDDLWPNPLNYFNSEDP 731
Query: 253 ----ASDGESGDLDDDDDDEDEEDDEEEVEVEKEEE 284
D E+GD DDDD +++DD+E+ + ++EEE
Sbjct: 732 DEAEEDDDEAGDAGKDDDDSEDDDDQEDDDDDEEEE 839
>TC89447 similar to PIR|S60892|S60892 nucleosome assembly protein 1 -
soybean, partial (34%)
Length = 803
Score = 90.5 bits (223), Expect = 7e-19
Identities = 45/76 (59%), Positives = 59/76 (77%)
Frame = +3
Query: 2 IQALTQQRYKALQYLFASPIVEKRLEVLKEIQKEHDDLKAKFFEERAALEAKYQLLYQPL 61
+Q L R ++ L SP V++R+EVLK +Q EHD+L++K+ EERA LEAKYQ LY+PL
Sbjct: 246 LQLLAGDRVDVMETL--SPNVKQRVEVLKVLQSEHDELESKYLEERAQLEAKYQKLYEPL 419
Query: 62 YTKRYEIVNGLAEVEG 77
YTKRYEIVNG+ +VEG
Sbjct: 420 YTKRYEIVNGVIDVEG 467
Score = 47.8 bits (112), Expect(2) = 5e-11
Identities = 19/33 (57%), Positives = 26/33 (78%)
Frame = +1
Query: 118 KDIKYTRTTEPQGFKLEFFFDSNPYFSNTILTK 150
KDIK+++ P+GFKL+F+FDSNPYF N+ K
Sbjct: 619 KDIKWSKLDNPKGFKLDFYFDSNPYFPNSRAXK 717
Score = 36.6 bits (83), Expect(2) = 5e-11
Identities = 15/32 (46%), Positives = 23/32 (71%)
Frame = +2
Query: 147 ILTKTYHMVDEDEFIMEKAIGTEIEWLPGKSL 178
+LTKTYHM+++D+ I+ K+ TEI+ P L
Sbjct: 707 VLTKTYHMIEDDDPILXKSXXTEIDXXPENCL 802
>TC91658 similar to GP|14329812|emb|CAC40753. putative nucleosome assembly
protein 1 {Atropa belladonna}, partial (46%)
Length = 583
Score = 74.7 bits (182), Expect = 4e-14
Identities = 35/73 (47%), Positives = 54/73 (73%), Gaps = 2/73 (2%)
Frame = +2
Query: 236 TIRDKIIPNAVSWYTGEASDGESGDL--DDDDDDEDEEDDEEEVEVEKEEEDDNYGLDDD 293
TIRDKIIP+AVSW+TGEA + + D+ D+DD+D DEEDD+++ + + +EED+ DD+
Sbjct: 2 TIRDKIIPHAVSWFTGEAGESDFDDIEVDEDDEDGDEEDDDDDDDDDDDEEDE----DDE 169
Query: 294 EEDDNEDVNNPKK 306
EED+ + + K+
Sbjct: 170 EEDEGKGKSKSKR 208
>TC77337 weakly similar to GP|4019275|gb|AAC95573.1| orf 48 {Ateline
herpesvirus 3}, partial (8%)
Length = 986
Score = 61.2 bits (147), Expect = 5e-10
Identities = 39/105 (37%), Positives = 56/105 (53%), Gaps = 8/105 (7%)
Frame = +3
Query: 211 DGMGVDEEADEELQNEMEHDYDIGSTIRD--KIIPNAVSW---YTGEASDGESGDLDDDD 265
DG G E ++EL +E Y S + K P + GE D E GD D+D
Sbjct: 456 DGDGAFGEGEDELSSEDGGGYGNNSNNKSNSKKAPEGGAGGADENGEEEDDEDGDDQDED 635
Query: 266 DDEDEEDDEEEVEVEKEEE---DDNYGLDDDEEDDNEDVNNPKKR 307
DDEDE+DD+EE E++EE D+ +++E++D E + PKKR
Sbjct: 636 DDEDEDDDDEEEGGEEDEEEGVDEEDNEEEEEDEDEEALQPPKKR 770
Score = 32.7 bits (73), Expect = 0.18
Identities = 25/81 (30%), Positives = 34/81 (41%), Gaps = 35/81 (43%)
Frame = +3
Query: 255 DGESGDLDDDDDDEDEEDD------EEEVEVEK--------------------------- 281
D E D +DD+D+ED++ D E+E+ E
Sbjct: 408 DAEGEDGNDDEDEEDDDGDGAFGEGEDELSSEDGGGYGNNSNNKSNSKKAPEGGAGGADE 587
Query: 282 --EEEDDNYGLDDDEEDDNED 300
EEEDD G DD +EDD+ED
Sbjct: 588 NGEEEDDEDG-DDQDEDDDED 647
Score = 27.3 bits (59), Expect = 7.6
Identities = 16/44 (36%), Positives = 22/44 (49%)
Frame = +3
Query: 254 SDGESGDLDDDDDDEDEEDDEEEVEVEKEEEDDNYGLDDDEEDD 297
S +S D D + ED DDE+E E+DD G + ED+
Sbjct: 378 SKTQSQDKQHDAEGEDGNDDEDE------EDDDGDGAFGEGEDE 491
>TC76514 homologue to PIR|T09648|T09648 nucleolin homolog nuM1 - alfalfa,
partial (61%)
Length = 1288
Score = 50.8 bits (120), Expect = 6e-07
Identities = 46/159 (28%), Positives = 75/159 (46%), Gaps = 6/159 (3%)
Frame = +3
Query: 156 DEDEFIMEKAIGTEIEWLPGKSL-TEKKDPNTAKQTIETEKFDSFFNFFNSLEIPKDGMG 214
+EDE KA+ ++ +P K TE D ++ E +K + + S K
Sbjct: 666 EEDEKPAAKAVPSKNGSVPAKKADTESSDEDSESSDEEDKKPAAKASKNVSAPTKKAASS 845
Query: 215 VDEEADEELQNEMEHDYDIGSTIRDKIIPNAVSWYTGEASDGESGDLDDDDDDEDEEDDE 274
DEE+DEE + D D + P AV+ + S +S D DD+DDD ++D+
Sbjct: 846 SDEESDEE----SDEDEDAKPVSK----PAAVA----KKSKKDSSDSDDEDDDSSSDEDK 989
Query: 275 EEVEVEKEEE--DDNYGLDDD---EEDDNEDVNNPKKRV 308
+ V +KE++ D G D D EE ++E P+K++
Sbjct: 990 KPVAAKKEDKMNVDKDGSDSDQSEEESEDEPSKTPQKKI 1106
Score = 34.7 bits (78), Expect = 0.048
Identities = 44/191 (23%), Positives = 73/191 (38%), Gaps = 38/191 (19%)
Frame = +3
Query: 156 DEDEFI--MEKAIGTEIEWLPGKSLTEKKDPNTAKQTIETEKFDSFFNFFNSLEIPKDGM 213
DE+E + KA+ ++ P K + ++ ++ + + E +K + + P
Sbjct: 444 DEEEVKKPVSKAVPSKNGSAPAKKVDTSEEEDSEESSDEDKKPAAKAVPSKNGSAPAKKA 623
Query: 214 GVDEEADEELQNEMEHDYDIGSTIRDKIIPNAVSWYTGEASDGESGDLDDDDDDE----- 268
DEE +E +E E D + K +P+ + +D ES D D + DE
Sbjct: 624 ASDEEDTDESSDEDEEDEKPAA----KAVPSKNGSVPAKKADTESSDEDSESSDEEDKKP 791
Query: 269 -------------------DEEDDEEEVE------------VEKEEEDDNYGLDDDEEDD 297
DEE DEE E V K+ + D+ D D+EDD
Sbjct: 792 AAKASKNVSAPTKKAASSSDEESDEESDEDEDAKPVSKPAAVAKKSKKDS--SDSDDEDD 965
Query: 298 NEDVNNPKKRV 308
+ + KK V
Sbjct: 966 DSSSDEDKKPV 998
>TC87688 similar to GP|10177535|dbj|BAB10930. gene_id:K1F13.21~unknown
protein {Arabidopsis thaliana}, partial (46%)
Length = 2077
Score = 50.4 bits (119), Expect = 8e-07
Identities = 33/99 (33%), Positives = 51/99 (51%), Gaps = 2/99 (2%)
Frame = +2
Query: 207 EIPKDGMGVDEEADEELQNEMEHDYDIGSTIRDKI--IPNAVSWYTGEASDGESGDLDDD 264
E +D G ++E DEE + E G I DK I + E + E G+ D+
Sbjct: 491 EDDEDEEGSEDEDDEEDEKEKVK----GGGIEDKFLKIDELTEYLEKEEDNYEKGEERDE 658
Query: 265 DDDEDEEDDEEEVEVEKEEEDDNYGLDDDEEDDNEDVNN 303
D++ EEDDE E E E +D++ DD+E+++ ED+ N
Sbjct: 659 ADEDSEEDDELEKAGEFEMDDEDDDDDDEEDEEAEDMGN 775
Score = 39.7 bits (91), Expect = 0.001
Identities = 25/63 (39%), Positives = 33/63 (51%), Gaps = 13/63 (20%)
Frame = +2
Query: 252 EASDGESGDLDDDDDD-------------EDEEDDEEEVEVEKEEEDDNYGLDDDEEDDN 298
E SD +LDDDDDD E E+D EEE + ++E +D +DDEED+
Sbjct: 380 EESDDFDEELDDDDDDFEGVEKKKAKGGSEGEDDFEEEDDEDEEGSED----EDDEEDEK 547
Query: 299 EDV 301
E V
Sbjct: 548 EKV 556
Score = 39.3 bits (90), Expect = 0.002
Identities = 30/117 (25%), Positives = 53/117 (44%), Gaps = 23/117 (19%)
Frame = +2
Query: 207 EIPKD-GMGVDEEADEELQNEMEHDYDIGSTIRDKIIPNAVSWYTGEASDGESGDLDDDD 265
++P D G ++++ EL+ E D+D D S+GE ++DD
Sbjct: 320 KVPLDVGKKLEKKKRVELEEEESDDFDEELDDDDDDFEGVEKKKAKGGSEGEDDFEEEDD 499
Query: 266 DDED--EEDDEEEVEVEK--------------------EEEDDNYGLDDDEEDDNED 300
+DE+ E++D+EE E EK E+E+DNY ++ ++ +ED
Sbjct: 500 EDEEGSEDEDDEEDEKEKVKGGGIEDKFLKIDELTEYLEKEEDNYEKGEERDEADED 670
Score = 35.8 bits (81), Expect = 0.021
Identities = 23/84 (27%), Positives = 44/84 (52%)
Frame = +2
Query: 217 EEADEELQNEMEHDYDIGSTIRDKIIPNAVSWYTGEASDGESGDLDDDDDDEDEEDDEEE 276
+E E L+ E E +Y+ G RD+ ++ E ++DD+DDD+D+E+DEE
Sbjct: 593 DELTEYLEKE-EDNYEKGEE-RDEADEDSEE--DDELEKAGEFEMDDEDDDDDDEEDEEA 760
Query: 277 VEVEKEEEDDNYGLDDDEEDDNED 300
++ +D +G +++ +D
Sbjct: 761 EDMGNVRYEDFFGGKNEKGSKRKD 832
Score = 29.3 bits (64), Expect = 2.0
Identities = 14/36 (38%), Positives = 22/36 (60%)
Frame = +2
Query: 272 DDEEEVEVEKEEEDDNYGLDDDEEDDNEDVNNPKKR 307
+ ++ VE+E+EE DD DD++DD E V K +
Sbjct: 350 EKKKRVELEEEESDDFDEELDDDDDDFEGVEKKKAK 457
>TC80893 similar to GP|4557063|gb|AAD22502.1| expressed protein {Arabidopsis
thaliana}, partial (39%)
Length = 883
Score = 47.0 bits (110), Expect = 9e-06
Identities = 28/75 (37%), Positives = 40/75 (53%), Gaps = 4/75 (5%)
Frame = +3
Query: 216 DEEADEELQNE----MEHDYDIGSTIRDKIIPNAVSWYTGEASDGESGDLDDDDDDEDEE 271
D++ D+++Q+E E DY G ++ P G + D DDD D+EDEE
Sbjct: 336 DDDDDDDVQDEDDDGEEEDYS-GDEGEEEGDPEDDPEANGAGGSDDGEDDDDDGDEEDEE 512
Query: 272 DDEEEVEVEKEEEDD 286
D E+E + E EEEDD
Sbjct: 513 DGEDEEDEEDEEEDD 557
Score = 43.5 bits (101), Expect = 1e-04
Identities = 31/98 (31%), Positives = 49/98 (49%), Gaps = 10/98 (10%)
Frame = +3
Query: 220 DEELQNEMEHDYDIGSTIRDKIIPNAVSWYTGEASDGESGDLDDD---------DDDEDE 270
+ + E + D D ++D+ Y+G+ + E GD +DD DD ED+
Sbjct: 306 ENKSDTEDDEDDDDDDDVQDEDDDGEEEDYSGDEGE-EEGDPEDDPEANGAGGSDDGEDD 482
Query: 271 EDD-EEEVEVEKEEEDDNYGLDDDEEDDNEDVNNPKKR 307
+DD +EE E + E+E+D +D+EEDD KKR
Sbjct: 483 DDDGDEEDEEDGEDEEDE---EDEEEDDETPQPPAKKR 587
Score = 43.1 bits (100), Expect = 1e-04
Identities = 25/64 (39%), Positives = 32/64 (49%), Gaps = 12/64 (18%)
Frame = +3
Query: 249 YTGEASDGESGDLDDDDDDEDEEDD------------EEEVEVEKEEEDDNYGLDDDEED 296
Y SD E + DDDDDD +EDD EEE + E + E + G DD ED
Sbjct: 300 YKENKSDTEDDEDDDDDDDVQDEDDDGEEEDYSGDEGEEEGDPEDDPEANGAGGSDDGED 479
Query: 297 DNED 300
D++D
Sbjct: 480 DDDD 491
Score = 32.3 bits (72), Expect = 0.24
Identities = 31/140 (22%), Positives = 59/140 (42%)
Frame = +3
Query: 144 SNTILTKTYHMVDEDEFIMEKAIGTEIEWLPGKSLTEKKDPNTAKQTIETEKFDSFFNFF 203
+N++ K M + F +E +G + + KS TE + + ++ E D +
Sbjct: 222 ANSVAPKLTAM--DSRFPLEHLLGRKPAYKENKSDTEDDEDDDDDDDVQDEDDDGEEEDY 395
Query: 204 NSLEIPKDGMGVDEEADEELQNEMEHDYDIGSTIRDKIIPNAVSWYTGEASDGESGDLDD 263
+ E ++G D E D E D G D G+ D E G+ ++
Sbjct: 396 SGDEGEEEG---DPEDDPEANGA--GGSDDGEDDDDD----------GDEEDEEDGEDEE 530
Query: 264 DDDDEDEEDDEEEVEVEKEE 283
D++DE+E+D+ + +K +
Sbjct: 531 DEEDEEEDDETPQPPAKKRK 590
>AW690594 similar to GP|23498163|emb hypothetical protein {Plasmodium
falciparum 3D7}, partial (10%)
Length = 633
Score = 47.0 bits (110), Expect = 9e-06
Identities = 27/88 (30%), Positives = 47/88 (52%)
Frame = +3
Query: 215 VDEEADEELQNEMEHDYDIGSTIRDKIIPNAVSWYTGEASDGESGDLDDDDDDEDEEDDE 274
V+E +EE + E+E + D E E + ++++++EDE DDE
Sbjct: 42 VEEHNEEEDEGEVEEEEDEHDEEE-------------EDEHDEEEEEEEEEEEEDEHDDE 182
Query: 275 EEVEVEKEEEDDNYGLDDDEEDDNEDVN 302
EE E E+EEE++ DDDEE + ++++
Sbjct: 183 EEEEEEEEEEEEE---DDDEEGEEDEID 257
Score = 43.1 bits (100), Expect = 1e-04
Identities = 18/56 (32%), Positives = 40/56 (71%)
Frame = +3
Query: 251 GEASDGESGDLDDDDDDEDEEDDEEEVEVEKEEEDDNYGLDDDEEDDNEDVNNPKK 306
GE + E + D++++DE +E++EEE E E+E+E D+ +++EE++ E+ ++ ++
Sbjct: 72 GEVEE-EEDEHDEEEEDEHDEEEEEEEEEEEEDEHDDEEEEEEEEEEEEEEDDDEE 236
Score = 37.0 bits (84), Expect = 0.010
Identities = 17/55 (30%), Positives = 34/55 (60%), Gaps = 1/55 (1%)
Frame = +3
Query: 247 SWYTGEASDGESGDLDDDDDDEDEE-DDEEEVEVEKEEEDDNYGLDDDEEDDNED 300
SW + + + + + ++E++E D+EEE E ++EEE++ ++DE DD E+
Sbjct: 24 SWKRRKVEEHNEEEDEGEVEEEEDEHDEEEEDEHDEEEEEEEEEEEEDEHDDEEE 188
>TC89427 weakly similar to GP|4557063|gb|AAD22502.1| expressed protein
{Arabidopsis thaliana}, partial (50%)
Length = 753
Score = 45.8 bits (107), Expect = 2e-05
Identities = 31/92 (33%), Positives = 45/92 (48%)
Frame = +1
Query: 216 DEEADEELQNEMEHDYDIGSTIRDKIIPNAVSWYTGEASDGESGDLDDDDDDEDEEDDEE 275
D++ DE+ + E D D D +PN G G DDD+DD+D++DD +
Sbjct: 355 DDDNDEDFSGD-EDDEDADP--EDDPVPN-----------GAGGSDDDDEDDDDDDDDND 492
Query: 276 EVEVEKEEEDDNYGLDDDEEDDNEDVNNPKKR 307
+ E E E D+EEDD+ED KK+
Sbjct: 493 DGEDEDE---------DEEEDDDEDQPPSKKK 561
Score = 43.5 bits (101), Expect = 1e-04
Identities = 21/65 (32%), Positives = 37/65 (56%), Gaps = 13/65 (20%)
Frame = +1
Query: 252 EASDGESGDLDDDDDDEDEEDDEEEVEVEKEEEDDNYGL-------------DDDEEDDN 298
+ S+ E D +DDDDD ++EDD+ + + +E+D++ DDDE+DD+
Sbjct: 295 DGSETEDDDDEDDDDDVNDEDDDNDEDFSGDEDDEDADPEDDPVPNGAGGSDDDDEDDDD 474
Query: 299 EDVNN 303
+D +N
Sbjct: 475 DDDDN 489
Score = 39.7 bits (91), Expect = 0.001
Identities = 16/44 (36%), Positives = 30/44 (67%)
Frame = +1
Query: 257 ESGDLDDDDDDEDEEDDEEEVEVEKEEEDDNYGLDDDEEDDNED 300
E+ D + +DD+DE+DD++ + + + ++D G +DDE+ D ED
Sbjct: 286 ENKDGSETEDDDDEDDDDDVNDEDDDNDEDFSGDEDDEDADPED 417
Score = 39.7 bits (91), Expect = 0.001
Identities = 22/65 (33%), Positives = 36/65 (54%), Gaps = 16/65 (24%)
Frame = +1
Query: 252 EASDGESGDLDDDDDDED----EEDDEEEVEVEKEEEDDNY------------GLDDDEE 295
E DG + DDDD+D+D +EDD+ + + +E+D++ G DDD+E
Sbjct: 286 ENKDGSETE-DDDDEDDDDDVNDEDDDNDEDFSGDEDDEDADPEDDPVPNGAGGSDDDDE 462
Query: 296 DDNED 300
DD++D
Sbjct: 463 DDDDD 477
Score = 28.9 bits (63), Expect = 2.6
Identities = 11/23 (47%), Positives = 18/23 (77%)
Frame = +1
Query: 281 KEEEDDNYGLDDDEEDDNEDVNN 303
+E +D + DDD+EDD++DVN+
Sbjct: 283 EENKDGSETEDDDDEDDDDDVND 351
>BF641220 weakly similar to PIR|G86203|G86 probable N-arginine dibasic
convertase [imported] - Arabidopsis thaliana, partial
(5%)
Length = 634
Score = 44.7 bits (104), Expect = 5e-05
Identities = 19/44 (43%), Positives = 32/44 (72%)
Frame = +2
Query: 257 ESGDLDDDDDDEDEEDDEEEVEVEKEEEDDNYGLDDDEEDDNED 300
+ G +D+DD++ED+ED+E++ EE+DD D+DEE ++ED
Sbjct: 344 KDGSIDEDDEEEDDEDEEDD-----EEDDDEGEDDEDEEXEDED 460
>TC93070 weakly similar to GP|17065230|gb|AAL32769.1 Unknown protein
{Arabidopsis thaliana}, partial (19%)
Length = 679
Score = 44.7 bits (104), Expect = 5e-05
Identities = 21/41 (51%), Positives = 31/41 (75%)
Frame = +1
Query: 260 DLDDDDDDEDEEDDEEEVEVEKEEEDDNYGLDDDEEDDNED 300
D DD+ ++ D EDDE+E E E E++DD+ DDD+EDD++D
Sbjct: 220 DSDDEIEEMDFEDDEDEDEDEDEDDDDDD--DDDDEDDDDD 336
Score = 37.7 bits (86), Expect = 0.006
Identities = 20/57 (35%), Positives = 35/57 (61%)
Frame = +1
Query: 252 EASDGESGDLDDDDDDEDEEDDEEEVEVEKEEEDDNYGLDDDEEDDNEDVNNPKKRV 308
E + + D +D+D+DEDE+DD+++ + ++EDD DDD + D+N KR+
Sbjct: 232 EIEEMDFEDDEDEDEDEDEDDDDDD---DDDDEDD----DDDGFAEPTDLNAKDKRL 381
>TC82080 similar to GP|9757854|dbj|BAB08488.1 contains similarity to
apoptosis antagonizing transcription
factor~gene_id:MFB13.10, partial (37%)
Length = 908
Score = 43.9 bits (102), Expect = 8e-05
Identities = 18/41 (43%), Positives = 30/41 (72%)
Frame = +2
Query: 260 DLDDDDDDEDEEDDEEEVEVEKEEEDDNYGLDDDEEDDNED 300
DLD+ DDD + ++ EVE+EEE+D+ G D++EED+ ++
Sbjct: 68 DLDEYDDDMSFNEVNDDSEVEEEEEEDDEGSDEEEEDERQE 190
Score = 32.3 bits (72), Expect = 0.24
Identities = 18/51 (35%), Positives = 27/51 (52%)
Frame = +2
Query: 249 YTGEASDGESGDLDDDDDDEDEEDDEEEVEVEKEEEDDNYGLDDDEEDDNE 299
Y + S E D D + ++E+EEDDE E E++E + DDE + E
Sbjct: 80 YDDDMSFNEVND-DSEVEEEEEEDDEGSDEEEEDERQEESRWKDDEMEQLE 229
Score = 29.3 bits (64), Expect = 2.0
Identities = 15/58 (25%), Positives = 31/58 (52%), Gaps = 8/58 (13%)
Frame = +2
Query: 252 EASDGESGDLDDD--------DDDEDEEDDEEEVEVEKEEEDDNYGLDDDEEDDNEDV 301
+ SD + + DDD D + +EE++E++ ++EEED+ ++D+ E +
Sbjct: 53 QKSDSDLDEYDDDMSFNEVNDDSEVEEEEEEDDEGSDEEEEDERQEESRWKDDEMEQL 226
Score = 26.9 bits (58), Expect = 9.9
Identities = 10/35 (28%), Positives = 22/35 (62%)
Frame = +2
Query: 262 DDDDDDEDEEDDEEEVEVEKEEEDDNYGLDDDEED 296
++DD+ DEE+++E E + ++D+ L+ + D
Sbjct: 140 EEDDEGSDEEEEDERQEESRWKDDEMEQLEKEYMD 244
Score = 26.9 bits (58), Expect = 9.9
Identities = 20/83 (24%), Positives = 36/83 (43%), Gaps = 2/83 (2%)
Frame = +2
Query: 205 SLEIPKDGMGVDEEADEELQNEMEHDYDIGSTIRDKIIPNAVSWYTGEASDGESGDLDDD 264
S ++ K +DE D+ NE+ D ++ E D E D +++
Sbjct: 41 SKKMQKSDSDLDEYDDDMSFNEVNDDSEVEEE---------------EEEDDEGSDEEEE 175
Query: 265 DDDEDEE--DDEEEVEVEKEEED 285
D+ ++E D+E ++EKE D
Sbjct: 176 DERQEESRWKDDEMEQLEKEYMD 244
>TC81365 similar to GP|14579399|gb|AAK69274.1 unknown {Glycine max}, partial
(38%)
Length = 920
Score = 43.5 bits (101), Expect = 1e-04
Identities = 20/53 (37%), Positives = 31/53 (57%)
Frame = +1
Query: 255 DGESGDLDDDDDDEDEEDDEEEVEVEKEEEDDNYGLDDDEEDDNEDVNNPKKR 307
+G G+ ++D+D++DE +D+ E E E+ E YG EED+N D K R
Sbjct: 274 NGGGGEEEEDEDEDDEWEDDSEEEEEESGEGGGYGSKGHEEDENIDHLRKKSR 432
>AW257087 homologue to GP|6683126|dbj KIAA0339 protein {Homo sapiens},
partial (2%)
Length = 466
Score = 43.5 bits (101), Expect = 1e-04
Identities = 24/57 (42%), Positives = 34/57 (59%), Gaps = 2/57 (3%)
Frame = -3
Query: 246 VSWYTGEASDGESGD--LDDDDDDEDEEDDEEEVEVEKEEEDDNYGLDDDEEDDNED 300
V + TG AS S LDD+D+DEDE++DE+ E E+EDD L++ E + D
Sbjct: 434 VFFTTGFASSSSSSSSLLDDEDEDEDEDEDED----EDEDEDDEELLEESESESESD 276
Score = 33.5 bits (75), Expect = 0.11
Identities = 14/33 (42%), Positives = 22/33 (66%)
Frame = -3
Query: 255 DGESGDLDDDDDDEDEEDDEEEVEVEKEEEDDN 287
D + + +D+D+DEDE+D+E E E E E D+
Sbjct: 371 DEDEDEDEDEDEDEDEDDEELLEESESESESDS 273
>TC80972 weakly similar to GP|23496972|gb|AAN36523.1 hypothetical protein
{Plasmodium falciparum 3D7}, partial (9%)
Length = 662
Score = 43.1 bits (100), Expect = 1e-04
Identities = 18/33 (54%), Positives = 26/33 (78%)
Frame = -2
Query: 257 ESGDLDDDDDDEDEEDDEEEVEVEKEEEDDNYG 289
E D DD DDD +E+D+EE+ EVE+EE+++ YG
Sbjct: 628 EEDDDDDADDDNEEKDEEEDDEVEEEEQEELYG 530
Score = 31.2 bits (69), Expect = 0.53
Identities = 13/35 (37%), Positives = 25/35 (71%)
Frame = -2
Query: 267 DEDEEDDEEEVEVEKEEEDDNYGLDDDEEDDNEDV 301
+ED++DD ++ EK+EE+D D+ EE++ E++
Sbjct: 628 EEDDDDDADDDNEEKDEEED----DEVEEEEQEEL 536
>TC80981 similar to GP|8698730|gb|AAF78488.1| ESTs gb|Z18526 gb|AI994480
gb|T44186 gb|Z30806 come from this gene. {Arabidopsis
thaliana}, partial (37%)
Length = 699
Score = 42.7 bits (99), Expect = 2e-04
Identities = 24/67 (35%), Positives = 35/67 (51%), Gaps = 18/67 (26%)
Frame = +3
Query: 254 SDGESGDLDDDDDDEDEEDDEEEVEV------------------EKEEEDDNYGLDDDEE 295
++ E+ DDD+DDEDEE+ +EE V E+E+EDD+ D D+
Sbjct: 231 NEPENNTDDDDEDDEDEENSDEEPVVDRKGKGIMRDDKGKGKMIEEEDEDDDDSDDSDDS 410
Query: 296 DDNEDVN 302
DD+ D N
Sbjct: 411 DDDSDGN 431
Score = 31.2 bits (69), Expect = 0.53
Identities = 22/78 (28%), Positives = 35/78 (44%), Gaps = 1/78 (1%)
Frame = +3
Query: 209 PKDGMGVDEEADEELQNEMEHDYDIGSTIRDKIIPNAVSWYTGEASDGESGDLDDDD-DD 267
P++ D+E DE+ +N E + D+ + G+ E D DDDD DD
Sbjct: 237 PENNTDDDDEDDEDEENSDEEP------VVDRKGKGIMRDDKGKGKMIEEEDEDDDDSDD 398
Query: 268 EDEEDDEEEVEVEKEEED 285
D+ DD+ + V + D
Sbjct: 399 SDDSDDDSDGNVSGSDSD 452
>CA859251 similar to GP|21305823|gb DNA polymerase I {Hz-1 insect virus}
[Heliothis zea virus 1], partial (2%)
Length = 803
Score = 42.7 bits (99), Expect = 2e-04
Identities = 25/56 (44%), Positives = 34/56 (60%), Gaps = 2/56 (3%)
Frame = -1
Query: 254 SDGESGDLDDDDDDEDEEDDEEEVEVEKEEEDD--NYGLDDDEEDDNEDVNNPKKR 307
SD +DDDDD+E+E++DE+E E E EEE+D NY +N D N+ KR
Sbjct: 671 SDKVRYKMDDDDDEEEEDEDEDENENEDEEEEDWLNY--------NNLDSNSHSKR 528
Score = 26.9 bits (58), Expect = 9.9
Identities = 13/41 (31%), Positives = 26/41 (62%), Gaps = 1/41 (2%)
Frame = -1
Query: 261 LDDDDDDEDEEDDEEEVEVEKEEEDDNYGLDDDE-EDDNED 300
+D + D + D+ ++V + +++DD D+DE E++NED
Sbjct: 710 IDMECVDVELVDNSDKVRYKMDDDDDEEEEDEDEDENENED 588
>TC76523 homologue to PIR|T09648|T09648 nucleolin homolog nuM1 - alfalfa,
partial (69%)
Length = 1615
Score = 42.7 bits (99), Expect = 2e-04
Identities = 26/82 (31%), Positives = 40/82 (48%)
Frame = +3
Query: 219 ADEELQNEMEHDYDIGSTIRDKIIPNAVSWYTGEASDGESGDLDDDDDDEDEEDDEEEVE 278
+DEE E + D D + P AV+ + S +S D DD+DDD ++D++ V
Sbjct: 168 SDEESDEESDEDEDAKPVSK----PAAVA----KKSKKDSSDSDDEDDDSSSDEDKKPVA 323
Query: 279 VEKEEEDDNYGLDDDEEDDNED 300
+KE + DDE+ N D
Sbjct: 324 AKKEVSESESDSSDDEDKMNVD 389
Score = 31.6 bits (70), Expect = 0.40
Identities = 15/48 (31%), Positives = 28/48 (58%)
Frame = +3
Query: 253 ASDGESGDLDDDDDDEDEEDDEEEVEVEKEEEDDNYGLDDDEEDDNED 300
+SD ES + + D+DED + + V K+ + D+ DD+++D + D
Sbjct: 165 SSDEESDE--ESDEDEDAKPVSKPAAVAKKSKKDSSDSDDEDDDSSSD 302
Score = 30.4 bits (67), Expect = 0.90
Identities = 15/56 (26%), Positives = 29/56 (51%)
Frame = +3
Query: 253 ASDGESGDLDDDDDDEDEEDDEEEVEVEKEEEDDNYGLDDDEEDDNEDVNNPKKRV 308
+S E D + D+D++ + + +K ++D + D D+EDD+ + KK V
Sbjct: 162 SSSDEESDEESDEDEDAKPVSKPAAVAKKSKKDSS---DSDDEDDDSSSDEDKKPV 320
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.310 0.134 0.374
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 7,490,839
Number of Sequences: 36976
Number of extensions: 95721
Number of successful extensions: 4813
Number of sequences better than 10.0: 377
Number of HSP's better than 10.0 without gapping: 1419
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 2869
length of query: 308
length of database: 9,014,727
effective HSP length: 96
effective length of query: 212
effective length of database: 5,465,031
effective search space: 1158586572
effective search space used: 1158586572
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.7 bits)
S2: 58 (26.9 bits)
Medicago: description of AC138526.4


