

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC138451.8 - phase: 0
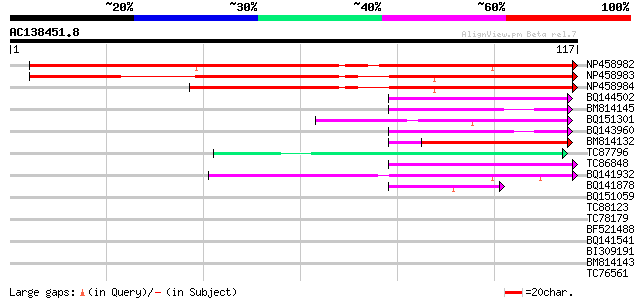
(117 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
NP458982 NP458982|AF498992.1|AAM18951.1 nodule-specific glycine-... 157 6e-40
NP458983 NP458983|AF498991.1|AAM18950.1 nodule-specific glycine-... 107 5e-25
NP458984 NP458984|AF498990.1|AAM18949.1 nodule-specific glycine-... 86 2e-18
BQ144502 weakly similar to GP|6523547|emb hydroxyproline-rich gl... 42 3e-05
BM814145 homologue to GP|21322711|em pherophorin-dz1 protein {Vo... 40 1e-04
BQ151301 similar to PIR|S34666|S346 glycine-rich protein - commo... 39 4e-04
BQ143960 weakly similar to SP|P21997|SSGP_ Sulfated surface glyc... 39 4e-04
BM814132 weakly similar to OMNI|MT3615.1 PE_PGRS family protein ... 38 5e-04
TC87796 weakly similar to SP|P03211|EBN1_EBV EBNA-1 nuclear prot... 38 6e-04
TC86848 similar to GP|10177413|dbj|BAB10544. fibrillarin-like {A... 38 6e-04
BQ141932 weakly similar to GP|6523547|emb hydroxyproline-rich gl... 38 6e-04
BQ141878 similar to GP|12697907|db KIAA1681 protein {Homo sapien... 37 8e-04
BQ151059 similar to GP|7291525|gb|A CG9888-PA {Drosophila melano... 37 0.001
TC88123 similar to GP|8777484|dbj|BAA97064.1 contains similarity... 37 0.001
TC78179 weakly similar to GP|7211427|gb|AAF40306.1| RNA helicase... 37 0.001
BF521488 weakly similar to GP|18447433|gb RE28280p {Drosophila m... 36 0.002
BQ141541 similar to GP|334068|gb|A ORF2 {Pseudorabies virus}, pa... 36 0.002
BI309191 35 0.003
BM814143 GP|21322711|e pherophorin-dz1 protein {Volvox carteri f... 35 0.003
TC76561 similar to PIR|S71779|S71779 glycine-rich RNA-binding pr... 35 0.003
>NP458982 NP458982|AF498992.1|AAM18951.1 nodule-specific glycine-rich protein
1C [Medicago truncatula]
Length = 447
Score = 157 bits (397), Expect = 6e-40
Identities = 80/118 (67%), Positives = 88/118 (73%), Gaps = 5/118 (4%)
Frame = +1
Query: 5 KKSKINIGVHEYANWLGKGTLLQEGDRNTNECG--GAILKGNEGKSNDGCGDGIEIHCRK 62
KKS INI VH Y NWL +GT+LQ GDRNTNE G GAI GN+G++NDGCGDGIE HC K
Sbjct: 88 KKSNINIEVHGYENWLSRGTILQGGDRNTNEFGYLGAIWNGNKGENNDGCGDGIEGHCGK 267
Query: 63 GYVDGTIWDKDISITKGTGWQTTGRKGGGSGKGGKG---GGGGYRIPIPGVGKGGGGK 117
GY+D T W KD T G GW +G KGGG GKGGKG GGGGY+IPIPG GKG GGK
Sbjct: 268 GYIDVT-WQKDH--TWGRGWWASGGKGGGGGKGGKGGKGGGGGYKIPIPGGGKGDGGK 432
>NP458983 NP458983|AF498991.1|AAM18950.1 nodule-specific glycine-rich protein
1B [Medicago truncatula]
Length = 393
Score = 107 bits (268), Expect = 5e-25
Identities = 61/115 (53%), Positives = 70/115 (60%), Gaps = 2/115 (1%)
Frame = +1
Query: 5 KKSKINIGVHEYANWLGKGTLLQEGDRNTNECGGAILKGNEGKSNDGCGDGIEIHCRKGY 64
KKSKINIGV+ YANWLG+G AI G +G++ DG DGIE HC KGY
Sbjct: 91 KKSKINIGVYGYANWLGRG---------------AIWNGKKGENIDGFVDGIEGHCGKGY 225
Query: 65 VDGTIWDKDISITKGTGWQTTG--RKGGGSGKGGKGGGGGYRIPIPGVGKGGGGK 117
VD T W K G GW T+ +GG GKGGKG GGGY+IP+P GKGGG K
Sbjct: 226 VDNT-WKK------GPGWGTSWWWGRGGKGGKGGKGSGGGYKIPLPVEGKGGGEK 369
>NP458984 NP458984|AF498990.1|AAM18949.1 nodule-specific glycine-rich protein
1A [Medicago truncatula]
Length = 327
Score = 85.9 bits (211), Expect = 2e-18
Identities = 46/82 (56%), Positives = 53/82 (64%), Gaps = 2/82 (2%)
Frame = +1
Query: 38 GAILKGNEGKSNDGCGDGIEIHCRKGYVDGTIWDKDISITKGTGWQTTG--RKGGGSGKG 95
GAI G +G++ DG DGIE HC KGYVD T W K G GW T+ +GG GKG
Sbjct: 79 GAIWNGKKGENIDGFVDGIEGHCGKGYVDNT-WKK------GPGWGTSWWWGRGGKGGKG 237
Query: 96 GKGGGGGYRIPIPGVGKGGGGK 117
GKG GGGY+IP+P GKGGG K
Sbjct: 238 GKGSGGGYKIPLPVEGKGGGEK 303
>BQ144502 weakly similar to GP|6523547|emb hydroxyproline-rich glycoprotein
DZ-HRGP {Volvox carteri f. nagariensis}, partial (39%)
Length = 1358
Score = 42.0 bits (97), Expect = 3e-05
Identities = 20/38 (52%), Positives = 21/38 (54%)
Frame = +3
Query: 79 GTGWQTTGRKGGGSGKGGKGGGGGYRIPIPGVGKGGGG 116
G+GW G GGG GG GGGGG R G GGGG
Sbjct: 87 GSGWCVWGGGGGGGRCGGGGGGGGGRGGAEAAGPGGGG 200
Score = 34.3 bits (77), Expect = 0.007
Identities = 20/51 (39%), Positives = 21/51 (40%), Gaps = 15/51 (29%)
Frame = +3
Query: 81 GWQTTGRKGG---------------GSGKGGKGGGGGYRIPIPGVGKGGGG 116
GW+ G GG G G GG GG GG PG G GGGG
Sbjct: 60 GWRRGGEVGGSGWCVWGGGGGGGRCGGGGGGGGGRGGAEAAGPGGGGGGGG 212
Score = 33.1 bits (74), Expect = 0.016
Identities = 18/39 (46%), Positives = 21/39 (53%), Gaps = 2/39 (5%)
Frame = +2
Query: 81 GWQTTG--RKGGGSGKGGKGGGGGYRIPIPGVGKGGGGK 117
GW G R G G G+G +GGGGG G GGGG+
Sbjct: 110 GWGRGGPLRGGWGGGRGPRGGGGG--------GPGGGGR 202
Score = 30.4 bits (67), Expect = 0.10
Identities = 25/83 (30%), Positives = 32/83 (38%), Gaps = 12/83 (14%)
Frame = +2
Query: 46 GKSNDGCGDGIEIHCRKGYVDGTIWDKDISITKGTGWQTTGRKGGGS----GKGGKGGGG 101
G+ G G G +G GT G G G + GG G+ G+ GG
Sbjct: 464 GRRGGGAGGGARPRIVEGLWGGT----------GAGGLAGGGEVGGGM*RLGREGRAPGG 613
Query: 102 GY--------RIPIPGVGKGGGG 116
R+PIPG +GGGG
Sbjct: 614 AAHQFSWQEGRVPIPGGEEGGGG 682
Score = 30.0 bits (66), Expect = 0.14
Identities = 15/32 (46%), Positives = 17/32 (52%)
Frame = +2
Query: 86 GRKGGGSGKGGKGGGGGYRIPIPGVGKGGGGK 117
GR GG G+G G GGG G G GGG +
Sbjct: 404 GRAGGRRGRGRWGAGGGRAGGRRGGGAGGGAR 499
Score = 28.5 bits (62), Expect = 0.39
Identities = 16/32 (50%), Positives = 17/32 (53%), Gaps = 4/32 (12%)
Frame = +1
Query: 87 RKGGGSGKGGKGGGGG----YRIPIPGVGKGG 114
+KGGG G G GGGGG GVG GG
Sbjct: 31 QKGGGRGGRGAGGGGGRWGAQAGACGGVGAGG 126
Score = 28.1 bits (61), Expect = 0.51
Identities = 27/79 (34%), Positives = 35/79 (44%), Gaps = 7/79 (8%)
Frame = +1
Query: 29 GDRNTNECGGAILKGNEGKSNDGCGDGIEIHCRKGYVD----GTIWDKD---ISITKGTG 81
G ++CGGA L G+ G G G G +GYV GT + I + +GT
Sbjct: 487 GGGAASDCGGA-LGGDGGGGTGGGGRG-----GRGYVATRTRGTSAGRGCPPIFLARGT- 645
Query: 82 WQTTGRKGGGSGKGGKGGG 100
R G G GG+GGG
Sbjct: 646 -----RSHSGGGGGGRGGG 687
Score = 28.1 bits (61), Expect = 0.51
Identities = 20/52 (38%), Positives = 22/52 (41%), Gaps = 15/52 (28%)
Frame = +2
Query: 79 GTGWQTTGRKGGGSGK-----GGKGGGGGYRIPIP----------GVGKGGG 115
G G G GGG G GG GGGG +R P G G+GGG
Sbjct: 116 GRGGPLRGGWGGGRGPRGGGGGGPGGGGRWRGGCPRPQAAC*GEWGRGRGGG 271
Score = 28.1 bits (61), Expect = 0.51
Identities = 19/42 (45%), Positives = 22/42 (52%), Gaps = 2/42 (4%)
Frame = +3
Query: 78 KGTGWQTTGRKGGGSGKG--GKGGGGGYRIPIPGVGKGGGGK 117
+G G G + GGSG G GGGGG G G GGGG+
Sbjct: 48 RGAGGWRRGGEVGGSGWCVWGGGGGGG---RCGGGGGGGGGR 164
Score = 27.3 bits (59), Expect = 0.88
Identities = 14/29 (48%), Positives = 17/29 (58%)
Frame = +2
Query: 86 GRKGGGSGKGGKGGGGGYRIPIPGVGKGG 114
G GGG GG GGGG + + G G+GG
Sbjct: 44 GAGGGGLAAGG-GGGGLRLVRVGGWGRGG 127
Score = 26.9 bits (58), Expect = 1.1
Identities = 15/38 (39%), Positives = 20/38 (52%)
Frame = +3
Query: 79 GTGWQTTGRKGGGSGKGGKGGGGGYRIPIPGVGKGGGG 116
G G G + G +G GG+GGG G G+ +G GG
Sbjct: 432 GAGGPEGGARAGAAG-GGRGGGRGL-----GLWRGSGG 527
Score = 26.2 bits (56), Expect = 2.0
Identities = 16/34 (47%), Positives = 17/34 (49%), Gaps = 3/34 (8%)
Frame = +1
Query: 86 GRKGG-GSGKGGKGGGGGYRIPIPGV--GKGGGG 116
GR+G G G GGGGG G G GGGG
Sbjct: 442 GRRGARGRAPRGGGGGGGAASDCGGALGGDGGGG 543
Score = 26.2 bits (56), Expect = 2.0
Identities = 17/47 (36%), Positives = 22/47 (46%), Gaps = 11/47 (23%)
Frame = +2
Query: 81 GWQTTGRKGGGSGK-----------GGKGGGGGYRIPIPGVGKGGGG 116
G + GR+GGG+G GG G GG + G G+ GGG
Sbjct: 449 GGRAGGRRGGGAGGGARPRIVEGLWGGTGAGG-----LAGGGEVGGG 574
Score = 26.2 bits (56), Expect = 2.0
Identities = 13/30 (43%), Positives = 15/30 (49%)
Frame = +1
Query: 88 KGGGSGKGGKGGGGGYRIPIPGVGKGGGGK 117
+GGG G G GG G G GGGG+
Sbjct: 472 RGGGGGGGAASDCGGALGGDGGGGTGGGGR 561
Score = 25.8 bits (55), Expect = 2.6
Identities = 18/51 (35%), Positives = 19/51 (36%), Gaps = 13/51 (25%)
Frame = +3
Query: 79 GTGWQTTGRKGGGSGKG-------------GKGGGGGYRIPIPGVGKGGGG 116
G G GGG G G G GGGGG R +GGGG
Sbjct: 159 GRGGAEAAGPGGGGGGGGDALGRRRLAKGSGAGGGGGAR------SRGGGG 293
Score = 25.8 bits (55), Expect = 2.6
Identities = 10/15 (66%), Positives = 11/15 (72%)
Frame = +2
Query: 90 GGSGKGGKGGGGGYR 104
GG GG GGGGG+R
Sbjct: 659 GGEEGGGGGGGGGWR 703
Score = 25.4 bits (54), Expect = 3.3
Identities = 11/19 (57%), Positives = 14/19 (72%)
Frame = +3
Query: 86 GRKGGGSGKGGKGGGGGYR 104
GR+G G G+ +GGGGG R
Sbjct: 663 GRRGAGGGE--EGGGGGVR 713
Score = 24.6 bits (52), Expect = 5.7
Identities = 16/42 (38%), Positives = 21/42 (49%), Gaps = 3/42 (7%)
Frame = +2
Query: 78 KGTG-WQTTGRKGGGSGKGGKGGGGGYRIPIPGV--GKGGGG 116
+G G W G + GG +GG GGG + G+ G G GG
Sbjct: 422 RGRGRWGAGGGRAGGR-RGGGAGGGARPRIVEGLWGGTGAGG 544
Score = 23.9 bits (50), Expect = 9.7
Identities = 15/35 (42%), Positives = 16/35 (44%), Gaps = 4/35 (11%)
Frame = +1
Query: 86 GRKGGGSGKGGKGGG----GGYRIPIPGVGKGGGG 116
G GG+ GG GGG GG R G G GG
Sbjct: 109 GVGAGGAAAGGVGGGAGAEGGRRRRARGGGAVAGG 213
>BM814145 homologue to GP|21322711|em pherophorin-dz1 protein {Volvox carteri
f. nagariensis}, partial (3%)
Length = 111
Score = 40.4 bits (93), Expect = 1e-04
Identities = 21/38 (55%), Positives = 21/38 (55%)
Frame = +3
Query: 79 GTGWQTTGRKGGGSGKGGKGGGGGYRIPIPGVGKGGGG 116
G GW G GGG G GG GGGGG G G GGGG
Sbjct: 12 GGGWGGGGGGGGGGGGGGGGGGGG------GGGGGGGG 107
Score = 36.2 bits (82), Expect = 0.002
Identities = 20/38 (52%), Positives = 20/38 (52%)
Frame = +2
Query: 79 GTGWQTTGRKGGGSGKGGKGGGGGYRIPIPGVGKGGGG 116
G G G GGG G GG GGGGG G G GGGG
Sbjct: 2 GGGGGGVGGGGGGGGGGGGGGGGG------GGGGGGGG 97
Score = 36.2 bits (82), Expect = 0.002
Identities = 20/38 (52%), Positives = 20/38 (52%)
Frame = +2
Query: 79 GTGWQTTGRKGGGSGKGGKGGGGGYRIPIPGVGKGGGG 116
G G G GGG G GG GGGGG G G GGGG
Sbjct: 5 GGGGGVGGGGGGGGGGGGGGGGGG------GGGGGGGG 100
Score = 31.2 bits (69), Expect = 0.061
Identities = 18/38 (47%), Positives = 18/38 (47%)
Frame = +1
Query: 79 GTGWQTTGRKGGGSGKGGKGGGGGYRIPIPGVGKGGGG 116
G G G GGG G GG GGGGG GGGG
Sbjct: 28 GGGGGGGGGGGGGGGGGGGGGGGG----------GGGG 111
>BQ151301 similar to PIR|S34666|S346 glycine-rich protein - common tobacco,
partial (22%)
Length = 530
Score = 38.5 bits (88), Expect = 4e-04
Identities = 24/57 (42%), Positives = 26/57 (45%), Gaps = 4/57 (7%)
Frame = -3
Query: 64 YVDGTIWDKDISITKGTGWQTTGRKGGGSGK----GGKGGGGGYRIPIPGVGKGGGG 116
+ G W+K G GW G GGG G GG GGGGG I P GGGG
Sbjct: 288 FFPGKGWEKKRKKRVGGGW--VGGVGGGGGGCHVGGGGGGGGGGIITAPASWGGGGG 124
Score = 36.6 bits (83), Expect = 0.001
Identities = 21/41 (51%), Positives = 23/41 (55%), Gaps = 3/41 (7%)
Frame = -1
Query: 79 GTGWQTTGRKGGGSG---KGGKGGGGGYRIPIPGVGKGGGG 116
G GW G GGG G +GG GG GG +P P G GGGG
Sbjct: 239 GGGWVVWG--GGGVGAMWEGGGGGAGGA*LPPPPHGGGGGG 123
Score = 29.6 bits (65), Expect = 0.18
Identities = 18/44 (40%), Positives = 22/44 (49%), Gaps = 5/44 (11%)
Frame = -2
Query: 78 KGTGWQTTGRKGGGSG-----KGGKGGGGGYRIPIPGVGKGGGG 116
K +G G GGG G +GG GG GG+ +G GGGG
Sbjct: 253 KKSGGGVGGWCGGGGGWVPCGRGGGGGRGGHNYRPRLMGGGGGG 122
Score = 25.4 bits (54), Expect = 3.3
Identities = 14/37 (37%), Positives = 18/37 (47%), Gaps = 11/37 (29%)
Frame = -1
Query: 92 SGKGGKGGGGG-----------YRIPIPGVGKGGGGK 117
+G+GG GGGGG +P P +G GGK
Sbjct: 374 NGRGGGGGGGGVFCFLFFLSFLLLLPPPPFFRGRGGK 264
Score = 23.9 bits (50), Expect = 9.7
Identities = 11/23 (47%), Positives = 13/23 (55%)
Frame = +2
Query: 86 GRKGGGSGKGGKGGGGGYRIPIP 108
G+KGGG G GK G + P P
Sbjct: 281 GKKGGGGGGEGKKERKGNKRPPP 349
>BQ143960 weakly similar to SP|P21997|SSGP_ Sulfated surface glycoprotein 185
(SSG 185). {Volvox carteri}, partial (13%)
Length = 1023
Score = 38.5 bits (88), Expect = 4e-04
Identities = 20/38 (52%), Positives = 23/38 (59%)
Frame = -2
Query: 79 GTGWQTTGRKGGGSGKGGKGGGGGYRIPIPGVGKGGGG 116
G G + GR+ GG GG+GGGGG R G GKGG G
Sbjct: 116 GEGGKENGREMGGK*GGGRGGGGGRR----GRGKGGAG 15
Score = 28.9 bits (63), Expect = 0.30
Identities = 14/26 (53%), Positives = 17/26 (64%), Gaps = 2/26 (7%)
Frame = -1
Query: 81 GWQTTGRKGGGSGKGGKG--GGGGYR 104
G + G +GGG G+ GKG GGGG R
Sbjct: 84 GGEMRGGEGGGGGEEGKGEGGGGGTR 7
Score = 26.6 bits (57), Expect = 1.5
Identities = 15/42 (35%), Positives = 19/42 (44%)
Frame = -3
Query: 76 ITKGTGWQTTGRKGGGSGKGGKGGGGGYRIPIPGVGKGGGGK 117
+ K W +K G G +GGGGG G GGGG+
Sbjct: 136 VKKERKWVRGEKKMGEKWGGNEGGGGG--------GGGGGGE 35
>BM814132 weakly similar to OMNI|MT3615.1 PE_PGRS family protein
{Mycobacterium tuberculosis CDC1551}, partial (7%)
Length = 164
Score = 38.1 bits (87), Expect = 5e-04
Identities = 21/38 (55%), Positives = 21/38 (55%)
Frame = +2
Query: 79 GTGWQTTGRKGGGSGKGGKGGGGGYRIPIPGVGKGGGG 116
G G G GGG G GG GGGGG R G G GGGG
Sbjct: 23 GGGGGGGGGGGGGGGGGGGGGGGGGRGGGGGGGGGGGG 136
Score = 38.1 bits (87), Expect = 5e-04
Identities = 20/38 (52%), Positives = 21/38 (54%)
Frame = +1
Query: 79 GTGWQTTGRKGGGSGKGGKGGGGGYRIPIPGVGKGGGG 116
G G G GGG G+GG GGGGG G G GGGG
Sbjct: 19 GGGGGGGGGGGGGGGRGGGGGGGGGGAGGGGGGGGGGG 132
Score = 37.4 bits (85), Expect = 8e-04
Identities = 20/38 (52%), Positives = 21/38 (54%)
Frame = +1
Query: 79 GTGWQTTGRKGGGSGKGGKGGGGGYRIPIPGVGKGGGG 116
G G G GGG G GG+GGGGG G G GGGG
Sbjct: 10 GGGGGGGGGGGGGGGGGGRGGGGGGGGGGAGGGGGGGG 123
Score = 37.4 bits (85), Expect = 8e-04
Identities = 18/31 (58%), Positives = 19/31 (61%)
Frame = +3
Query: 86 GRKGGGSGKGGKGGGGGYRIPIPGVGKGGGG 116
G +GGG G GG GGGGG G G GGGG
Sbjct: 6 GGRGGGGGGGGGGGGGGGGAGGGGGGGGGGG 98
Score = 36.2 bits (82), Expect = 0.002
Identities = 20/38 (52%), Positives = 20/38 (52%)
Frame = +2
Query: 79 GTGWQTTGRKGGGSGKGGKGGGGGYRIPIPGVGKGGGG 116
G G GR GGG G GG GGGGG G GGGG
Sbjct: 74 GGGGGGGGRGGGGGGGGGGGGGGG--------GGGGGG 163
Score = 36.2 bits (82), Expect = 0.002
Identities = 20/38 (52%), Positives = 20/38 (52%)
Frame = +3
Query: 79 GTGWQTTGRKGGGSGKGGKGGGGGYRIPIPGVGKGGGG 116
G G G GGG G GG GGGGG G G GGGG
Sbjct: 60 GAGGGGGGGGGGGGGGGGGGGGGG------GGGGGGGG 155
Score = 36.2 bits (82), Expect = 0.002
Identities = 20/38 (52%), Positives = 20/38 (52%)
Frame = +3
Query: 79 GTGWQTTGRKGGGSGKGGKGGGGGYRIPIPGVGKGGGG 116
G G G GGG G GG GGGGG G G GGGG
Sbjct: 48 GGGGGAGGGGGGGGGGGGGGGGGG------GGGGGGGG 143
Score = 36.2 bits (82), Expect = 0.002
Identities = 20/38 (52%), Positives = 20/38 (52%)
Frame = +3
Query: 79 GTGWQTTGRKGGGSGKGGKGGGGGYRIPIPGVGKGGGG 116
G G G GGG G GG GGGGG G G GGGG
Sbjct: 45 GGGGGGAGGGGGGGGGGGGGGGGG------GGGGGGGG 140
Score = 36.2 bits (82), Expect = 0.002
Identities = 20/38 (52%), Positives = 21/38 (54%)
Frame = +2
Query: 79 GTGWQTTGRKGGGSGKGGKGGGGGYRIPIPGVGKGGGG 116
G G G GGG G GG GGGGG G G+GGGG
Sbjct: 8 GAGGGGGGGGGGGGGGGGGGGGGGGG---GGGGRGGGG 112
Score = 35.8 bits (81), Expect = 0.002
Identities = 18/31 (58%), Positives = 19/31 (61%)
Frame = +1
Query: 86 GRKGGGSGKGGKGGGGGYRIPIPGVGKGGGG 116
G GGG G GG GGGGG G G+GGGG
Sbjct: 1 GGGGGGGGGGGGGGGGG-----GGGGRGGGG 78
Score = 35.8 bits (81), Expect = 0.002
Identities = 20/39 (51%), Positives = 21/39 (53%)
Frame = +2
Query: 79 GTGWQTTGRKGGGSGKGGKGGGGGYRIPIPGVGKGGGGK 117
G G G GGG G GG GGGGG G G GGGG+
Sbjct: 2 GGGAGGGGGGGGGGGGGGGGGGGG------GGGGGGGGR 100
Score = 35.0 bits (79), Expect = 0.004
Identities = 19/38 (50%), Positives = 20/38 (52%)
Frame = +2
Query: 79 GTGWQTTGRKGGGSGKGGKGGGGGYRIPIPGVGKGGGG 116
G G G +GGG G GG GGGGG G GGGG
Sbjct: 71 GGGGGGGGGRGGGGGGGGGGGGGG--------GGGGGG 160
Score = 33.9 bits (76), Expect = 0.009
Identities = 19/39 (48%), Positives = 20/39 (50%)
Frame = +3
Query: 78 KGTGWQTTGRKGGGSGKGGKGGGGGYRIPIPGVGKGGGG 116
+G G G GGG G G GGGGG G G GGGG
Sbjct: 12 RGGGGGGGGGGGGGGGGAGGGGGGG------GGGGGGGG 110
Score = 33.5 bits (75), Expect = 0.012
Identities = 19/38 (50%), Positives = 19/38 (50%)
Frame = +1
Query: 79 GTGWQTTGRKGGGSGKGGKGGGGGYRIPIPGVGKGGGG 116
G G G GGG G G GGGGG G G GGGG
Sbjct: 13 GGGGGGGGGGGGGGGGGRGGGGGGGGGGAGGGGGGGGG 126
Score = 33.5 bits (75), Expect = 0.012
Identities = 19/38 (50%), Positives = 19/38 (50%)
Frame = +1
Query: 79 GTGWQTTGRKGGGSGKGGKGGGGGYRIPIPGVGKGGGG 116
G G G GGG G GG G GGG G G GGGG
Sbjct: 4 GGGGGGGGGGGGGGGGGGGGRGGGGGGGGGGAGGGGGG 117
>TC87796 weakly similar to SP|P03211|EBN1_EBV EBNA-1 nuclear protein.
[strain B95-8 Human herpesvirus 4] {Epstein-barr
virus}, partial (23%)
Length = 431
Score = 37.7 bits (86), Expect = 6e-04
Identities = 27/73 (36%), Positives = 29/73 (38%)
Frame = +1
Query: 43 GNEGKSNDGCGDGIEIHCRKGYVDGTIWDKDISITKGTGWQTTGRKGGGSGKGGKGGGGG 102
G G S G G G G G + G G + G GGG G GG GGGGG
Sbjct: 25 GGGGGSGGGGGGGA------GGAHGVGYGSGGGTGGGYGGGSPGGGGGGGGSGGGGGGGG 186
Query: 103 YRIPIPGVGKGGG 115
G G GGG
Sbjct: 187 AHGGAYGGGIGGG 225
Score = 34.7 bits (78), Expect = 0.006
Identities = 20/38 (52%), Positives = 20/38 (52%)
Frame = +1
Query: 79 GTGWQTTGRKGGGSGKGGKGGGGGYRIPIPGVGKGGGG 116
G G G G G G GG G GGGY PG G GGGG
Sbjct: 49 GGGGGAGGAHGVGYGSGG-GTGGGYGGGSPGGGGGGGG 159
Score = 33.5 bits (75), Expect = 0.012
Identities = 16/30 (53%), Positives = 16/30 (53%)
Frame = +1
Query: 86 GRKGGGSGKGGKGGGGGYRIPIPGVGKGGG 115
G GGG G GG GGGG G G GGG
Sbjct: 16 GGYGGGGGSGGGGGGGAGGAHGVGYGSGGG 105
Score = 30.0 bits (66), Expect = 0.14
Identities = 17/30 (56%), Positives = 17/30 (56%), Gaps = 2/30 (6%)
Frame = +1
Query: 89 GGGSGKGG--KGGGGGYRIPIPGVGKGGGG 116
GGG G GG GGGGG GVG G GG
Sbjct: 13 GGGYGGGGGSGGGGGGGAGGAHGVGYGSGG 102
Score = 26.6 bits (57), Expect = 1.5
Identities = 13/25 (52%), Positives = 13/25 (52%)
Frame = +1
Query: 79 GTGWQTTGRKGGGSGKGGKGGGGGY 103
G G G GGG G G GG GGY
Sbjct: 175 GGGGAHGGAYGGGIGGGEGGGHGGY 249
>TC86848 similar to GP|10177413|dbj|BAB10544. fibrillarin-like {Arabidopsis
thaliana}, partial (92%)
Length = 1250
Score = 37.7 bits (86), Expect = 6e-04
Identities = 18/39 (46%), Positives = 21/39 (53%)
Frame = +3
Query: 79 GTGWQTTGRKGGGSGKGGKGGGGGYRIPIPGVGKGGGGK 117
G G GR GG G GG+GG G G G+GGGG+
Sbjct: 102 GRGGDRGGRGGGRGGFGGRGGDRGTPFKARGGGRGGGGR 218
Score = 36.6 bits (83), Expect = 0.001
Identities = 29/81 (35%), Positives = 37/81 (44%)
Frame = +3
Query: 37 GGAILKGNEGKSNDGCGDGIEIHCRKGYVDGTIWDKDISITKGTGWQTTGRKGGGSGKGG 96
GG +G G G G G +G G D+ GT ++ +GGG G GG
Sbjct: 81 GGGGFRGGRGGDRGGRGGG------RGGFGGRGGDR------GTPFKA---RGGGRGGGG 215
Query: 97 KGGGGGYRIPIPGVGKGGGGK 117
+GG GG R G G+GGG K
Sbjct: 216 RGGRGGGR----GGGRGGGMK 266
Score = 28.1 bits (61), Expect = 0.51
Identities = 14/29 (48%), Positives = 18/29 (61%)
Frame = +3
Query: 88 KGGGSGKGGKGGGGGYRIPIPGVGKGGGG 116
+GGG +GG+GG G R G G+GG G
Sbjct: 78 RGGGGFRGGRGGDRGGR----GGGRGGFG 152
>BQ141932 weakly similar to GP|6523547|emb hydroxyproline-rich glycoprotein
DZ-HRGP {Volvox carteri f. nagariensis}, partial (30%)
Length = 1338
Score = 37.7 bits (86), Expect = 6e-04
Identities = 29/90 (32%), Positives = 39/90 (43%), Gaps = 14/90 (15%)
Frame = +1
Query: 42 KGNEGKSNDGCGDGIEIHCRKGYVDGTIWDKDISITKGTGWQTTGRKGGGSGKGGKG--- 98
+G EG G G G + + +G +D S G GW+ +G GGG G GG
Sbjct: 163 RGGEGGGWGGGGGGGGLVRGRRKEEGAWGGRDESW--GGGWRGSGGGGGGDGGGGNPAMG 336
Query: 99 --GGGGYRIPIP---------GVGKGGGGK 117
GGGG +P+ G GGGG+
Sbjct: 337 TKGGGGVELPMRRGGEGR*PWATGAGGGGR 426
Score = 31.2 bits (69), Expect = 0.061
Identities = 26/81 (32%), Positives = 33/81 (40%)
Frame = +2
Query: 36 CGGAILKGNEGKSNDGCGDGIEIHCRKGYVDGTIWDKDISITKGTGWQTTGRKGGGSGKG 95
CGG G G G G+G +G++ G G + GR GG + G
Sbjct: 155 CGGG---GGRGGGGGGEGEG------EGWLGG-------------GGKRKGRGGGETRAG 268
Query: 96 GKGGGGGYRIPIPGVGKGGGG 116
G+ GG GVG GGGG
Sbjct: 269 GEDGG--------GVGGGGGG 307
Score = 30.8 bits (68), Expect = 0.079
Identities = 29/104 (27%), Positives = 37/104 (34%), Gaps = 1/104 (0%)
Frame = +1
Query: 15 EYANWLGKGTLLQEGDRNTNECGGAILKGNEGKSNDGCGDGIEIHCRKGYVDGTIWDKDI 74
E W G+ G R + GG G G G+E+ R+G W
Sbjct: 232 EEGAWGGRDESWGGGWRGSGGGGGGDGGGGNPAMGTKGGGGVELPMRRGGEGR*PW---- 399
Query: 75 SITKGTGWQTTGRKGGGSGKGGKGGGGGYRIPIPG-VGKGGGGK 117
TG GGG GG+ GG R + G G GG G+
Sbjct: 400 ---------ATGAGGGGRRVGGRRGGERRRAWVGGRTGWGGAGR 504
Score = 30.4 bits (67), Expect = 0.10
Identities = 18/33 (54%), Positives = 19/33 (57%), Gaps = 1/33 (3%)
Frame = +2
Query: 86 GRKGGGSGKGGKGGGGGYRIPIPGVG-KGGGGK 117
G GGG G+GG GGG G G G GGGGK
Sbjct: 149 GGCGGGGGRGGGGGGEG-----EGEGWLGGGGK 232
Score = 27.3 bits (59), Expect = 0.88
Identities = 17/46 (36%), Positives = 21/46 (44%)
Frame = +2
Query: 70 WDKDISITKGTGWQTTGRKGGGSGKGGKGGGGGYRIPIPGVGKGGG 115
W D G G + G G G G+G GGGG + G+GGG
Sbjct: 134 WCLDWGGCGGGGGRGGGGGGEGEGEGWLGGGGKRK------GRGGG 253
Score = 25.8 bits (55), Expect = 2.6
Identities = 29/101 (28%), Positives = 36/101 (34%), Gaps = 10/101 (9%)
Frame = +2
Query: 12 GVHEYANWLGKGTLLQEGDRNTNECGGAILKGNEGKSNDGCGDGI---------EIHCRK 62
G E WLG G + GG G G G GI E +CR
Sbjct: 191 GEGEGEGWLGGGGKRKGRGGGETRAGGEDGGGVGGGGGGTAGGGIPQWVQRAEGEWNCRC 370
Query: 63 GYVDGTIWDKDISITKGTGWQTTG-RKGGGSGKGGKGGGGG 102
G + D+ G G + G GGG +G +G GGG
Sbjct: 371 GG-GARVDDRG---RPGRGGEVGGWGGGGGESEGARGWGGG 481
>BQ141878 similar to GP|12697907|db KIAA1681 protein {Homo sapiens}, partial
(2%)
Length = 1102
Score = 37.4 bits (85), Expect = 8e-04
Identities = 17/30 (56%), Positives = 18/30 (59%), Gaps = 6/30 (20%)
Frame = -2
Query: 79 GTGWQTTGRKGG------GSGKGGKGGGGG 102
GTGW T G GG G G+GG GGGGG
Sbjct: 90 GTGWSTAGPMGGRGG*GLGEGRGGGGGGGG 1
Score = 29.3 bits (64), Expect = 0.23
Identities = 17/40 (42%), Positives = 19/40 (47%), Gaps = 4/40 (10%)
Frame = -3
Query: 80 TGWQTTGRK----GGGSGKGGKGGGGGYRIPIPGVGKGGG 115
T W GR+ GGG G+G GGGG G GGG
Sbjct: 98 TSWGPVGRRRGRWGGGGGRGWVRGGGG--------GGGGG 3
>BQ151059 similar to GP|7291525|gb|A CG9888-PA {Drosophila melanogaster},
partial (26%)
Length = 308
Score = 37.0 bits (84), Expect = 0.001
Identities = 21/40 (52%), Positives = 21/40 (52%)
Frame = +3
Query: 77 TKGTGWQTTGRKGGGSGKGGKGGGGGYRIPIPGVGKGGGG 116
T G G G GGG G GG GGGGG G G GGGG
Sbjct: 30 TPGGGGGGGGGGGGGGGGGGGGGGGG------GGGGGGGG 131
Score = 36.6 bits (83), Expect = 0.001
Identities = 19/34 (55%), Positives = 20/34 (57%)
Frame = +3
Query: 83 QTTGRKGGGSGKGGKGGGGGYRIPIPGVGKGGGG 116
+T G GGG G GG GGGGG G G GGGG
Sbjct: 27 KTPGGGGGGGGGGGGGGGGG------GGGGGGGG 110
Score = 36.2 bits (82), Expect = 0.002
Identities = 20/39 (51%), Positives = 21/39 (53%)
Frame = +1
Query: 78 KGTGWQTTGRKGGGSGKGGKGGGGGYRIPIPGVGKGGGG 116
+G G G GGG G GG GGGGG G G GGGG
Sbjct: 34 RGGGGGGGGGGGGGGGGGGGGGGGG------GGGGGGGG 132
Score = 35.4 bits (80), Expect = 0.003
Identities = 20/38 (52%), Positives = 20/38 (52%)
Frame = +2
Query: 79 GTGWQTTGRKGGGSGKGGKGGGGGYRIPIPGVGKGGGG 116
G G G GGG G GG GGGGG G G GGGG
Sbjct: 41 GGGGGGGGGGGGGGGGGGGGGGGG------GGGGGGGG 136
Score = 35.4 bits (80), Expect = 0.003
Identities = 20/38 (52%), Positives = 20/38 (52%)
Frame = +3
Query: 79 GTGWQTTGRKGGGSGKGGKGGGGGYRIPIPGVGKGGGG 116
G G G GGG G GG GGGGG G G GGGG
Sbjct: 63 GGGGGGGGGGGGGGGGGGGGGGGG------GGGGGGGG 158
Score = 35.4 bits (80), Expect = 0.003
Identities = 20/38 (52%), Positives = 20/38 (52%)
Frame = +1
Query: 79 GTGWQTTGRKGGGSGKGGKGGGGGYRIPIPGVGKGGGG 116
G G G GGG G GG GGGGG G G GGGG
Sbjct: 64 GGGGGGGGGGGGGGGGGGGGGGGG------GGGGGGGG 159
Score = 35.4 bits (80), Expect = 0.003
Identities = 20/38 (52%), Positives = 20/38 (52%)
Frame = +2
Query: 79 GTGWQTTGRKGGGSGKGGKGGGGGYRIPIPGVGKGGGG 116
G G G GGG G GG GGGGG G G GGGG
Sbjct: 53 GGGGGGGGGGGGGGGGGGGGGGGG------GGGGGGGG 148
Score = 35.4 bits (80), Expect = 0.003
Identities = 20/38 (52%), Positives = 20/38 (52%)
Frame = +2
Query: 79 GTGWQTTGRKGGGSGKGGKGGGGGYRIPIPGVGKGGGG 116
G G G GGG G GG GGGGG G G GGGG
Sbjct: 47 GGGGGGGGGGGGGGGGGGGGGGGG------GGGGGGGG 142
Score = 35.4 bits (80), Expect = 0.003
Identities = 20/38 (52%), Positives = 20/38 (52%)
Frame = +3
Query: 79 GTGWQTTGRKGGGSGKGGKGGGGGYRIPIPGVGKGGGG 116
G G G GGG G GG GGGGG G G GGGG
Sbjct: 78 GGGGGGGGGGGGGGGGGGGGGGGG------GGGGGGGG 173
Score = 35.4 bits (80), Expect = 0.003
Identities = 20/38 (52%), Positives = 20/38 (52%)
Frame = +2
Query: 79 GTGWQTTGRKGGGSGKGGKGGGGGYRIPIPGVGKGGGG 116
G G G GGG G GG GGGGG G G GGGG
Sbjct: 80 GGGGGGGGGGGGGGGGGGGGGGGG------GGGGGGGG 175
Score = 35.4 bits (80), Expect = 0.003
Identities = 20/38 (52%), Positives = 20/38 (52%)
Frame = +2
Query: 79 GTGWQTTGRKGGGSGKGGKGGGGGYRIPIPGVGKGGGG 116
G G G GGG G GG GGGGG G G GGGG
Sbjct: 83 GGGGGGGGGGGGGGGGGGGGGGGG------GGGGGGGG 178
Score = 35.4 bits (80), Expect = 0.003
Identities = 20/38 (52%), Positives = 20/38 (52%)
Frame = +3
Query: 79 GTGWQTTGRKGGGSGKGGKGGGGGYRIPIPGVGKGGGG 116
G G G GGG G GG GGGGG G G GGGG
Sbjct: 87 GGGGGGGGGGGGGGGGGGGGGGGG------GGGGGGGG 182
Score = 35.4 bits (80), Expect = 0.003
Identities = 20/38 (52%), Positives = 20/38 (52%)
Frame = +3
Query: 79 GTGWQTTGRKGGGSGKGGKGGGGGYRIPIPGVGKGGGG 116
G G G GGG G GG GGGGG G G GGGG
Sbjct: 90 GGGGGGGGGGGGGGGGGGGGGGGG------GGGGGGGG 185
Score = 35.4 bits (80), Expect = 0.003
Identities = 20/38 (52%), Positives = 20/38 (52%)
Frame = +3
Query: 79 GTGWQTTGRKGGGSGKGGKGGGGGYRIPIPGVGKGGGG 116
G G G GGG G GG GGGGG G G GGGG
Sbjct: 93 GGGGGGGGGGGGGGGGGGGGGGGG------GGGGGGGG 188
Score = 35.4 bits (80), Expect = 0.003
Identities = 20/38 (52%), Positives = 20/38 (52%)
Frame = +1
Query: 79 GTGWQTTGRKGGGSGKGGKGGGGGYRIPIPGVGKGGGG 116
G G G GGG G GG GGGGG G G GGGG
Sbjct: 100 GGGGGGGGGGGGGGGGGGGGGGGG------GGGGGGGG 195
Score = 35.4 bits (80), Expect = 0.003
Identities = 20/38 (52%), Positives = 20/38 (52%)
Frame = +1
Query: 79 GTGWQTTGRKGGGSGKGGKGGGGGYRIPIPGVGKGGGG 116
G G G GGG G GG GGGGG G G GGGG
Sbjct: 199 GGGGGGGGGGGGGGGGGGGGGGGG------GGGGGGGG 294
Score = 35.4 bits (80), Expect = 0.003
Identities = 20/38 (52%), Positives = 20/38 (52%)
Frame = +1
Query: 79 GTGWQTTGRKGGGSGKGGKGGGGGYRIPIPGVGKGGGG 116
G G G GGG G GG GGGGG G G GGGG
Sbjct: 73 GGGGGGGGGGGGGGGGGGGGGGGG------GGGGGGGG 168
Score = 35.4 bits (80), Expect = 0.003
Identities = 20/38 (52%), Positives = 20/38 (52%)
Frame = +3
Query: 79 GTGWQTTGRKGGGSGKGGKGGGGGYRIPIPGVGKGGGG 116
G G G GGG G GG GGGGG G G GGGG
Sbjct: 114 GGGGGGGGGGGGGGGGGGGGGGGG------GGGGGGGG 209
Score = 35.4 bits (80), Expect = 0.003
Identities = 20/38 (52%), Positives = 20/38 (52%)
Frame = +3
Query: 79 GTGWQTTGRKGGGSGKGGKGGGGGYRIPIPGVGKGGGG 116
G G G GGG G GG GGGGG G G GGGG
Sbjct: 117 GGGGGGGGGGGGGGGGGGGGGGGG------GGGGGGGG 212
Score = 35.4 bits (80), Expect = 0.003
Identities = 20/38 (52%), Positives = 20/38 (52%)
Frame = +2
Query: 79 GTGWQTTGRKGGGSGKGGKGGGGGYRIPIPGVGKGGGG 116
G G G GGG G GG GGGGG G G GGGG
Sbjct: 125 GGGGGGGGGGGGGGGGGGGGGGGG------GGGGGGGG 220
Score = 35.4 bits (80), Expect = 0.003
Identities = 20/38 (52%), Positives = 20/38 (52%)
Frame = +3
Query: 79 GTGWQTTGRKGGGSGKGGKGGGGGYRIPIPGVGKGGGG 116
G G G GGG G GG GGGGG G G GGGG
Sbjct: 132 GGGGGGGGGGGGGGGGGGGGGGGG------GGGGGGGG 227
Score = 35.4 bits (80), Expect = 0.003
Identities = 20/38 (52%), Positives = 20/38 (52%)
Frame = +3
Query: 79 GTGWQTTGRKGGGSGKGGKGGGGGYRIPIPGVGKGGGG 116
G G G GGG G GG GGGGG G G GGGG
Sbjct: 138 GGGGGGGGGGGGGGGGGGGGGGGG------GGGGGGGG 233
Score = 35.4 bits (80), Expect = 0.003
Identities = 20/38 (52%), Positives = 20/38 (52%)
Frame = +1
Query: 79 GTGWQTTGRKGGGSGKGGKGGGGGYRIPIPGVGKGGGG 116
G G G GGG G GG GGGGG G G GGGG
Sbjct: 145 GGGGGGGGGGGGGGGGGGGGGGGG------GGGGGGGG 240
Score = 35.4 bits (80), Expect = 0.003
Identities = 20/38 (52%), Positives = 20/38 (52%)
Frame = +2
Query: 79 GTGWQTTGRKGGGSGKGGKGGGGGYRIPIPGVGKGGGG 116
G G G GGG G GG GGGGG G G GGGG
Sbjct: 149 GGGGGGGGGGGGGGGGGGGGGGGG------GGGGGGGG 244
Score = 35.4 bits (80), Expect = 0.003
Identities = 20/38 (52%), Positives = 20/38 (52%)
Frame = +3
Query: 79 GTGWQTTGRKGGGSGKGGKGGGGGYRIPIPGVGKGGGG 116
G G G GGG G GG GGGGG G G GGGG
Sbjct: 165 GGGGGGGGGGGGGGGGGGGGGGGG------GGGGGGGG 260
Score = 35.4 bits (80), Expect = 0.003
Identities = 20/38 (52%), Positives = 20/38 (52%)
Frame = +3
Query: 79 GTGWQTTGRKGGGSGKGGKGGGGGYRIPIPGVGKGGGG 116
G G G GGG G GG GGGGG G G GGGG
Sbjct: 168 GGGGGGGGGGGGGGGGGGGGGGGG------GGGGGGGG 263
Score = 35.4 bits (80), Expect = 0.003
Identities = 20/38 (52%), Positives = 20/38 (52%)
Frame = +1
Query: 79 GTGWQTTGRKGGGSGKGGKGGGGGYRIPIPGVGKGGGG 116
G G G GGG G GG GGGGG G G GGGG
Sbjct: 172 GGGGGGGGGGGGGGGGGGGGGGGG------GGGGGGGG 267
Score = 35.4 bits (80), Expect = 0.003
Identities = 20/38 (52%), Positives = 20/38 (52%)
Frame = +2
Query: 79 GTGWQTTGRKGGGSGKGGKGGGGGYRIPIPGVGKGGGG 116
G G G GGG G GG GGGGG G G GGGG
Sbjct: 176 GGGGGGGGGGGGGGGGGGGGGGGG------GGGGGGGG 271
Score = 35.4 bits (80), Expect = 0.003
Identities = 20/38 (52%), Positives = 20/38 (52%)
Frame = +3
Query: 79 GTGWQTTGRKGGGSGKGGKGGGGGYRIPIPGVGKGGGG 116
G G G GGG G GG GGGGG G G GGGG
Sbjct: 180 GGGGGGGGGGGGGGGGGGGGGGGG------GGGGGGGG 275
Score = 35.4 bits (80), Expect = 0.003
Identities = 20/38 (52%), Positives = 20/38 (52%)
Frame = +2
Query: 79 GTGWQTTGRKGGGSGKGGKGGGGGYRIPIPGVGKGGGG 116
G G G GGG G GG GGGGG G G GGGG
Sbjct: 185 GGGGGGGGGGGGGGGGGGGGGGGG------GGGGGGGG 280
Score = 35.4 bits (80), Expect = 0.003
Identities = 20/38 (52%), Positives = 20/38 (52%)
Frame = +3
Query: 79 GTGWQTTGRKGGGSGKGGKGGGGGYRIPIPGVGKGGGG 116
G G G GGG G GG GGGGG G G GGGG
Sbjct: 192 GGGGGGGGGGGGGGGGGGGGGGGG------GGGGGGGG 287
Score = 35.4 bits (80), Expect = 0.003
Identities = 20/38 (52%), Positives = 20/38 (52%)
Frame = +2
Query: 79 GTGWQTTGRKGGGSGKGGKGGGGGYRIPIPGVGKGGGG 116
G G G GGG G GG GGGGG G G GGGG
Sbjct: 98 GGGGGGGGGGGGGGGGGGGGGGGG------GGGGGGGG 193
Score = 35.4 bits (80), Expect = 0.003
Identities = 20/38 (52%), Positives = 20/38 (52%)
Frame = +1
Query: 79 GTGWQTTGRKGGGSGKGGKGGGGGYRIPIPGVGKGGGG 116
G G G GGG G GG GGGGG G G GGGG
Sbjct: 208 GGGGGGGGGGGGGGGGGGGGGGGG------GGGGGGGG 303
Score = 35.4 bits (80), Expect = 0.003
Identities = 20/38 (52%), Positives = 20/38 (52%)
Frame = +1
Query: 79 GTGWQTTGRKGGGSGKGGKGGGGGYRIPIPGVGKGGGG 116
G G G GGG G GG GGGGG G G GGGG
Sbjct: 211 GGGGGGGGGGGGGGGGGGGGGGGG------GGGGGGGG 306
Score = 35.4 bits (80), Expect = 0.003
Identities = 20/38 (52%), Positives = 20/38 (52%)
Frame = +1
Query: 79 GTGWQTTGRKGGGSGKGGKGGGGGYRIPIPGVGKGGGG 116
G G G GGG G GG GGGGG G G GGGG
Sbjct: 103 GGGGGGGGGGGGGGGGGGGGGGGG------GGGGGGGG 198
Score = 35.4 bits (80), Expect = 0.003
Identities = 20/38 (52%), Positives = 20/38 (52%)
Frame = +1
Query: 79 GTGWQTTGRKGGGSGKGGKGGGGGYRIPIPGVGKGGGG 116
G G G GGG G GG GGGGG G G GGGG
Sbjct: 67 GGGGGGGGGGGGGGGGGGGGGGGG------GGGGGGGG 162
Score = 35.0 bits (79), Expect = 0.004
Identities = 19/37 (51%), Positives = 20/37 (53%)
Frame = +2
Query: 80 TGWQTTGRKGGGSGKGGKGGGGGYRIPIPGVGKGGGG 116
+G G GGG G GG GGGGG G G GGGG
Sbjct: 20 SGENPGGGGGGGGGGGGGGGGGG------GGGGGGGG 112
>TC88123 similar to GP|8777484|dbj|BAA97064.1 contains similarity to
RNA-binding protein~gene_id:K15M2.15 {Arabidopsis
thaliana}, partial (11%)
Length = 815
Score = 36.6 bits (83), Expect = 0.001
Identities = 18/37 (48%), Positives = 19/37 (50%)
Frame = +1
Query: 79 GTGWQTTGRKGGGSGKGGKGGGGGYRIPIPGVGKGGG 115
G T GG G GG G GGG + PGVG GGG
Sbjct: 97 GYSGPTAAGYGGSGGGGGGGYGGGSGVTGPGVGAGGG 207
Score = 29.3 bits (64), Expect = 0.23
Identities = 21/51 (41%), Positives = 25/51 (48%), Gaps = 8/51 (15%)
Frame = +1
Query: 74 ISITKGTGWQTTGRKGGGS--GKGGKGGG--GGYRIPIP----GVGKGGGG 116
+S + G G + G + S G GG GGG GGY P G G GGGG
Sbjct: 1 LSSSVGGGLSSVGNQAPSSIGGAGGYGGGHYGGYSGPTAAGYGGSGGGGGG 153
>TC78179 weakly similar to GP|7211427|gb|AAF40306.1| RNA helicase {Vigna
radiata}, partial (12%)
Length = 692
Score = 36.6 bits (83), Expect = 0.001
Identities = 20/36 (55%), Positives = 24/36 (66%), Gaps = 1/36 (2%)
Frame = +2
Query: 83 QTTGRK-GGGSGKGGKGGGGGYRIPIPGVGKGGGGK 117
+++GR+ GGG G G GGGGYR G G GGGGK
Sbjct: 215 ESSGRRFGGGRGFSGGRGGGGYRFGGKG-GGGGGGK 319
Score = 33.1 bits (74), Expect = 0.016
Identities = 21/44 (47%), Positives = 25/44 (56%), Gaps = 5/44 (11%)
Frame = +2
Query: 79 GTGWQTTGRKGGG----SGKGGKGGGG-GYRIPIPGVGKGGGGK 117
G G +G +GGG GKGG GGGG GY G G+GGG +
Sbjct: 236 GGGRGFSGGRGGGGYRFGGKGGGGGGGKGY-----GGGRGGGNR 352
>BF521488 weakly similar to GP|18447433|gb RE28280p {Drosophila
melanogaster}, partial (84%)
Length = 470
Score = 35.8 bits (81), Expect = 0.002
Identities = 19/38 (50%), Positives = 20/38 (52%)
Frame = +2
Query: 79 GTGWQTTGRKGGGSGKGGKGGGGGYRIPIPGVGKGGGG 116
G G+ G GGG GG GGGG P G G GGGG
Sbjct: 209 GGGYNPYGGFGGGRPYGGFGGGGFGGRPFRGGGGGGGG 322
Score = 28.5 bits (62), Expect = 0.39
Identities = 18/40 (45%), Positives = 21/40 (52%), Gaps = 4/40 (10%)
Frame = +2
Query: 81 GWQTTGRKGGGSGKGGKGGG----GGYRIPIPGVGKGGGG 116
G+ G GGG G+ G GGG GG+ P G GGGG
Sbjct: 158 GFGGGGFGGGGFGRPGFGGGYNPYGGFGGGRPYGGFGGGG 277
Score = 27.7 bits (60), Expect = 0.67
Identities = 17/42 (40%), Positives = 18/42 (42%), Gaps = 4/42 (9%)
Frame = +2
Query: 79 GTGWQTTGRKGGGSG----KGGKGGGGGYRIPIPGVGKGGGG 116
G G G GGG G +GG GGGGG GGG
Sbjct: 239 GGGRPYGGFGGGGFGGRPFRGGGGGGGGSASASASANAAGGG 364
Score = 26.2 bits (56), Expect = 2.0
Identities = 14/25 (56%), Positives = 15/25 (60%), Gaps = 1/25 (4%)
Frame = +2
Query: 93 GKGGKGGGGGYRIP-IPGVGKGGGG 116
G GG G GGG+ P G G GGGG
Sbjct: 116 GFGGPGFGGGFGRPGFGGGGFGGGG 190
Score = 26.2 bits (56), Expect = 2.0
Identities = 17/38 (44%), Positives = 19/38 (49%), Gaps = 9/38 (23%)
Frame = +2
Query: 89 GGGSGKGGKGGG--GGYRIPIPGVGKG-------GGGK 117
GGG G+ G GGG GG PG G G GGG+
Sbjct: 137 GGGFGRPGFGGGGFGGGGFGRPGFGGGYNPYGGFGGGR 250
>BQ141541 similar to GP|334068|gb|A ORF2 {Pseudorabies virus}, partial (2%)
Length = 1320
Score = 35.8 bits (81), Expect = 0.002
Identities = 20/40 (50%), Positives = 22/40 (55%), Gaps = 9/40 (22%)
Frame = +3
Query: 86 GRKGGGSGKGGKGG---------GGGYRIPIPGVGKGGGG 116
GR+GGG G GG+GG GGG RI G K GGG
Sbjct: 591 GRRGGG*GGGGRGGAGAKRRRSRGGGGRIATAGSRKKGGG 710
Score = 24.6 bits (52), Expect = 5.7
Identities = 16/43 (37%), Positives = 18/43 (41%), Gaps = 8/43 (18%)
Frame = +2
Query: 67 GTIWDKDISITKGTG-WQTTGR-------KGGGSGKGGKGGGG 101
G W D KG G +T GR +G G GG GGG
Sbjct: 479 GRKWTSDAKKMKGAGRGETEGRGSIRKKRRGAGRRFGGAAGGG 607
>BI309191
Length = 109
Score = 35.4 bits (80), Expect = 0.003
Identities = 20/38 (52%), Positives = 20/38 (52%)
Frame = +3
Query: 79 GTGWQTTGRKGGGSGKGGKGGGGGYRIPIPGVGKGGGG 116
G G G GGG G GG GGGGG G G GGGG
Sbjct: 3 GGGGGGGGGGGGGGGGGGGGGGGG------GGGGGGGG 98
Score = 35.4 bits (80), Expect = 0.003
Identities = 20/38 (52%), Positives = 20/38 (52%)
Frame = +2
Query: 79 GTGWQTTGRKGGGSGKGGKGGGGGYRIPIPGVGKGGGG 116
G G G GGG G GG GGGGG G G GGGG
Sbjct: 11 GGGGGGGGGGGGGGGGGGGGGGGG------GGGGGGGG 106
Score = 35.4 bits (80), Expect = 0.003
Identities = 20/38 (52%), Positives = 20/38 (52%)
Frame = +2
Query: 79 GTGWQTTGRKGGGSGKGGKGGGGGYRIPIPGVGKGGGG 116
G G G GGG G GG GGGGG G G GGGG
Sbjct: 5 GGGGGGGGGGGGGGGGGGGGGGGG------GGGGGGGG 100
Score = 30.0 bits (66), Expect = 0.14
Identities = 14/24 (58%), Positives = 14/24 (58%)
Frame = +1
Query: 79 GTGWQTTGRKGGGSGKGGKGGGGG 102
G G G GGG G GG GGGGG
Sbjct: 37 GGGGGGGGGGGGGGGGGGGGGGGG 108
>BM814143 GP|21322711|e pherophorin-dz1 protein {Volvox carteri f.
nagariensis}, partial (4%)
Length = 132
Score = 35.4 bits (80), Expect = 0.003
Identities = 20/38 (52%), Positives = 20/38 (52%)
Frame = +1
Query: 79 GTGWQTTGRKGGGSGKGGKGGGGGYRIPIPGVGKGGGG 116
G G G GGG G GG GGGGG G G GGGG
Sbjct: 13 GGGGGGGGGGGGGGGGGGGGGGGG------GGGGGGGG 108
Score = 35.4 bits (80), Expect = 0.003
Identities = 20/38 (52%), Positives = 20/38 (52%)
Frame = +1
Query: 79 GTGWQTTGRKGGGSGKGGKGGGGGYRIPIPGVGKGGGG 116
G G G GGG G GG GGGGG G G GGGG
Sbjct: 10 GGGGGGGGGGGGGGGGGGGGGGGG------GGGGGGGG 105
Score = 35.4 bits (80), Expect = 0.003
Identities = 20/38 (52%), Positives = 20/38 (52%)
Frame = +3
Query: 79 GTGWQTTGRKGGGSGKGGKGGGGGYRIPIPGVGKGGGG 116
G G G GGG G GG GGGGG G G GGGG
Sbjct: 3 GGGGGGGGGGGGGGGGGGGGGGGG------GGGGGGGG 98
Score = 35.4 bits (80), Expect = 0.003
Identities = 20/38 (52%), Positives = 20/38 (52%)
Frame = +3
Query: 79 GTGWQTTGRKGGGSGKGGKGGGGGYRIPIPGVGKGGGG 116
G G G GGG G GG GGGGG G G GGGG
Sbjct: 18 GGGGGGGGGGGGGGGGGGGGGGGG------GGGGGGGG 113
Score = 35.4 bits (80), Expect = 0.003
Identities = 20/38 (52%), Positives = 20/38 (52%)
Frame = +2
Query: 79 GTGWQTTGRKGGGSGKGGKGGGGGYRIPIPGVGKGGGG 116
G G G GGG G GG GGGGG G G GGGG
Sbjct: 29 GGGGGGGGGGGGGGGGGGGGGGGG------GGGGGGGG 124
Score = 35.4 bits (80), Expect = 0.003
Identities = 20/38 (52%), Positives = 20/38 (52%)
Frame = +2
Query: 79 GTGWQTTGRKGGGSGKGGKGGGGGYRIPIPGVGKGGGG 116
G G G GGG G GG GGGGG G G GGGG
Sbjct: 32 GGGGGGGGGGGGGGGGGGGGGGGG------GGGGGGGG 127
Score = 35.4 bits (80), Expect = 0.003
Identities = 20/38 (52%), Positives = 20/38 (52%)
Frame = +3
Query: 79 GTGWQTTGRKGGGSGKGGKGGGGGYRIPIPGVGKGGGG 116
G G G GGG G GG GGGGG G G GGGG
Sbjct: 9 GGGGGGGGGGGGGGGGGGGGGGGG------GGGGGGGG 104
Score = 33.5 bits (75), Expect = 0.012
Identities = 19/38 (50%), Positives = 19/38 (50%)
Frame = +1
Query: 79 GTGWQTTGRKGGGSGKGGKGGGGGYRIPIPGVGKGGGG 116
G G G GGG G GG GGGGG G GGGG
Sbjct: 40 GGGGGGGGGGGGGGGGGGGGGGGG--------GGGGGG 129
Score = 30.0 bits (66), Expect = 0.14
Identities = 14/24 (58%), Positives = 14/24 (58%)
Frame = +1
Query: 79 GTGWQTTGRKGGGSGKGGKGGGGG 102
G G G GGG G GG GGGGG
Sbjct: 61 GGGGGGGGGGGGGGGGGGGGGGGG 132
>TC76561 similar to PIR|S71779|S71779 glycine-rich RNA-binding protein GRP1
- wheat, partial (95%)
Length = 1054
Score = 35.4 bits (80), Expect = 0.003
Identities = 31/96 (32%), Positives = 39/96 (40%)
Frame = +1
Query: 21 GKGTLLQEGDRNTNECGGAILKGNEGKSNDGCGDGIEIHCRKGYVDGTIWDKDISITKGT 80
G+ + + + GG +G G G G G E R+GY G
Sbjct: 559 GRNITVNQAQSRGSGGGGGGGRGGGGYGGGGGGYGGE---RRGY--------------GG 687
Query: 81 GWQTTGRKGGGSGKGGKGGGGGYRIPIPGVGKGGGG 116
G G GGG G+ +GGGGGY G G GGGG
Sbjct: 688 GGGYGGGGGGGYGER-RGGGGGYSRGGGGGGYGGGG 792
Score = 33.9 bits (76), Expect = 0.009
Identities = 27/80 (33%), Positives = 35/80 (43%), Gaps = 1/80 (1%)
Frame = +1
Query: 38 GAILKGNEGKSNDGCGDGIEIHCRKGY-VDGTIWDKDISITKGTGWQTTGRKGGGSGKGG 96
G + NE ND I G +DG + + ++G+G G +GGG GG
Sbjct: 487 GFVTFANEKSMNDA------IEAMNGQDLDGRNITVNQAQSRGSGGGGGGGRGGGGYGGG 648
Query: 97 KGGGGGYRIPIPGVGKGGGG 116
GG GG R G G GGG
Sbjct: 649 GGGYGGERRGYGGGGGYGGG 708
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.311 0.143 0.454
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 3,825,143
Number of Sequences: 36976
Number of extensions: 63204
Number of successful extensions: 2828
Number of sequences better than 10.0: 407
Number of HSP's better than 10.0 without gapping: 1011
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1737
length of query: 117
length of database: 9,014,727
effective HSP length: 93
effective length of query: 24
effective length of database: 5,575,959
effective search space: 133823016
effective search space used: 133823016
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.7 bits)
S2: 50 (23.9 bits)
Medicago: description of AC138451.8


