

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC137824.11 - phase: 0
(316 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
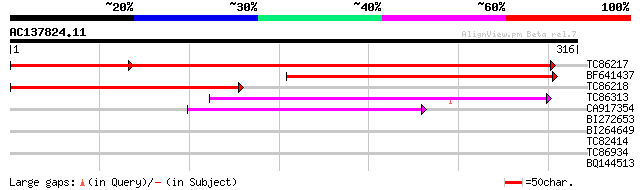
Score E
Sequences producing significant alignments: (bits) Value
TC86217 similar to GP|14488275|dbj|BAB60898. serine palmitoyltra... 559 e-160
BF641437 similar to GP|21450450|gb Putative serine palmitoyltran... 281 3e-76
TC86218 weakly similar to GP|14488275|dbj|BAB60898. serine palmi... 234 2e-62
TC86313 similar to GP|4995890|emb|CAB44316.1 serine palmitoyltra... 95 4e-20
CA917354 GP|490225|emb BIOE {Escherichia coli}, partial (46%) 64 6e-11
BI272653 weakly similar to PIR|E84871|E848 probable polygalactur... 30 1.2
BI264649 28 6.0
TC82414 similar to PIR|T49915|T49915 pre-mRNA splicing factor AT... 27 7.9
TC86934 similar to PIR|T04630|T04630 synaptobrevin homolog F10N7... 27 7.9
BQ144513 similar to GP|995935|gb|AA MAZ {Homo sapiens}, partial ... 27 7.9
>TC86217 similar to GP|14488275|dbj|BAB60898. serine palmitoyltransferase
{Arabidopsis thaliana}, partial (98%)
Length = 2027
Score = 559 bits (1441), Expect = e-160
Identities = 274/304 (90%), Positives = 290/304 (95%)
Frame = +2
Query: 1 MASSFINFVNTTIDLVTYALHAPSARAVVFGFNIGGHLFIEVLLLVVILFLLSQKSYKPP 60
MAS+ +NFVNTT++ VTYAL APSARA+VFG+N GGHLFIEV LLVVILFLLSQKSYKPP
Sbjct: 374 MASAAVNFVNTTLNWVTYALDAPSARAIVFGYNFGGHLFIEVFLLVVILFLLSQKSYKPP 553
Query: 61 KRPLSNKEIDELCDEWVPQPLIPSLNDEMPYEPPVLESAAGPHTIVNGKEVVNFASANYL 120
KRPL+NKEIDELCDEWVPQ LIPSL+ +M YEPPVLESAAGPHTIVNGKEV+NFASANYL
Sbjct: 554 KRPLTNKEIDELCDEWVPQSLIPSLDKDMHYEPPVLESAAGPHTIVNGKEVINFASANYL 733
Query: 121 GLIGHQKLLDSCSSALEKYGVGSCGPRGFYGTIDVHLDCEGRISKFLGTPDSILYSYGLS 180
GLIGHQKLLDSC+SALEKYGVGSCGPRGFYGTIDVHLDCE RI+KFLGTPDSILYSYGLS
Sbjct: 734 GLIGHQKLLDSCTSALEKYGVGSCGPRGFYGTIDVHLDCEARIAKFLGTPDSILYSYGLS 913
Query: 181 TMFSAIPAFSKKGDIIVADEGVHWGIQNGLYLSRSTVVYFKHNNMDSLRETLENITSIYK 240
TMFSAIPAFSKKGDIIVADEGVHWGIQNGLYLSRSTVVYFKHN+MDSL ETLE +TS YK
Sbjct: 914 TMFSAIPAFSKKGDIIVADEGVHWGIQNGLYLSRSTVVYFKHNDMDSLEETLEKLTSKYK 1093
Query: 241 RTKNLRRYIVIEALYQNSGQIAPLDEIIKLKEKYCFRILLDESNSFGVLGSSGRGLTEHY 300
TKNLRRYIV+EALYQNSGQIAPLD+IIKLKEKY FRILLDESNSFGVLGSSGRGLTEHY
Sbjct: 1094HTKNLRRYIVVEALYQNSGQIAPLDDIIKLKEKYRFRILLDESNSFGVLGSSGRGLTEHY 1273
Query: 301 GVPV 304
GVPV
Sbjct: 1274GVPV 1285
Score = 117 bits (294), Expect = 4e-27
Identities = 57/69 (82%), Positives = 64/69 (92%)
Frame = -3
Query: 1 MASSFINFVNTTIDLVTYALHAPSARAVVFGFNIGGHLFIEVLLLVVILFLLSQKSYKPP 60
MAS+ +NFVNTT++ VTYAL APSARA+VFG+N GGHLFIEV LLVVILFLLSQKSYKPP
Sbjct: 207 MASAAVNFVNTTLNWVTYALDAPSARAIVFGYNFGGHLFIEVFLLVVILFLLSQKSYKPP 28
Query: 61 KRPLSNKEI 69
KRPL+NKEI
Sbjct: 27 KRPLTNKEI 1
>BF641437 similar to GP|21450450|gb Putative serine palmitoyltransferase
{Oryza sativa (japonica cultivar-group)}, partial (37%)
Length = 655
Score = 281 bits (718), Expect = 3e-76
Identities = 138/151 (91%), Positives = 144/151 (94%)
Frame = +1
Query: 155 VHLDCEGRISKFLGTPDSILYSYGLSTMFSAIPAFSKKGDIIVADEGVHWGIQNGLYLSR 214
VHLDCE RI+KFL TPDSILYSYGLSTMFSAIPAFSKKGDIIVADEGVHWGIQNGLYLSR
Sbjct: 1 VHLDCEARIAKFLRTPDSILYSYGLSTMFSAIPAFSKKGDIIVADEGVHWGIQNGLYLSR 180
Query: 215 STVVYFKHNNMDSLRETLENITSIYKRTKNLRRYIVIEALYQNSGQIAPLDEIIKLKEKY 274
STVVYFKHN+MDSL ETLE ++S YK TKNLRRYIV+EALYQNSGQIAPLD+IIKLKEKY
Sbjct: 181 STVVYFKHNDMDSLEETLEKLSSKYKHTKNLRRYIVVEALYQNSGQIAPLDDIIKLKEKY 360
Query: 275 CFRILLDESNSFGVLGSSGRGLTEHYGVPVC 305
FRILLDESNSFGVLGSSGRGLTEHYGVPVC
Sbjct: 361 RFRILLDESNSFGVLGSSGRGLTEHYGVPVC 453
>TC86218 weakly similar to GP|14488275|dbj|BAB60898. serine
palmitoyltransferase {Arabidopsis thaliana}, partial
(26%)
Length = 997
Score = 234 bits (598), Expect = 2e-62
Identities = 113/130 (86%), Positives = 123/130 (93%)
Frame = +3
Query: 1 MASSFINFVNTTIDLVTYALHAPSARAVVFGFNIGGHLFIEVLLLVVILFLLSQKSYKPP 60
MAS+ +NFVNTT++ VTYAL APSARA+VFG+N GGHLFIEV LLVVILFLLSQKSYKPP
Sbjct: 606 MASAAVNFVNTTLNWVTYALDAPSARAIVFGYNFGGHLFIEVFLLVVILFLLSQKSYKPP 785
Query: 61 KRPLSNKEIDELCDEWVPQPLIPSLNDEMPYEPPVLESAAGPHTIVNGKEVVNFASANYL 120
KRPL+NKEIDELCDEWVPQPLIPSL+ +M YEPPVLESAAGPHTIVNGKEV+NFASANYL
Sbjct: 786 KRPLTNKEIDELCDEWVPQPLIPSLDKDMHYEPPVLESAAGPHTIVNGKEVINFASANYL 965
Query: 121 GLIGHQKLLD 130
GLIGHQKLLD
Sbjct: 966 GLIGHQKLLD 995
>TC86313 similar to GP|4995890|emb|CAB44316.1 serine palmitoyltransferase
{Solanum tuberosum}, partial (98%)
Length = 2052
Score = 94.7 bits (234), Expect = 4e-20
Identities = 60/195 (30%), Positives = 96/195 (48%), Gaps = 4/195 (2%)
Frame = +2
Query: 112 VNFASANYLGLIGHQKLLDS-CSSALEKYGVGSCGPRGFYGTIDVHLDCEGRISKFLGTP 170
+N S NYLG + L+KY +C R GT +H + E ++ F+ P
Sbjct: 521 LNLGSYNYLGFAAADEYCTPRVIDTLKKYSPSTCSTRVDGGTTALHNELEECVASFVKKP 700
Query: 171 DSILYSYGLSTMFSAIPAFSKKGDIIVADEGVHWGIQNGLYLSRSTVVYFKHNNMDSLRE 230
++++ G T + +P KG +I++D H I NG S +T+ F+HN L E
Sbjct: 701 AALVFGMGYVTNSAILPVLMGKGSLIISDSLNHNSIVNGARGSGATIRVFQHNVPSHLEE 880
Query: 231 TL-ENITSIYKRTKN--LRRYIVIEALYQNSGQIAPLDEIIKLKEKYCFRILLDESNSFG 287
L E I RT + +++E +Y G++ L E+I + +KY LDE++S G
Sbjct: 881 VLREQIAEGQPRTHRPWKKIMVIVEGIYSMEGELCKLPEVIAICKKYKAYTYLDEAHSIG 1060
Query: 288 VLGSSGRGLTEHYGV 302
+G +GRG+ E GV
Sbjct: 1061AVGKTGRGVCELLGV 1105
>CA917354 GP|490225|emb BIOE {Escherichia coli}, partial (46%)
Length = 676
Score = 64.3 bits (155), Expect = 6e-11
Identities = 39/133 (29%), Positives = 66/133 (49%)
Frame = -3
Query: 100 AGPHTIVNGKEVVNFASANYLGLIGHQKLLDSCSSALEKYGVGSCGPRGFYGTIDVHLDC 159
AG + + ++ +NF+S +YLGL H +++ + E++G+GS G G VH
Sbjct: 449 AGRWLVADDRQYLNFSSNDYLGLSHHPQIIRAWQQGAEQFGIGSGGSGHVSGYSVVHQAL 270
Query: 160 EGRISKFLGTPDSILYSYGLSTMFSAIPAFSKKGDIIVADEGVHWGIQNGLYLSRSTVVY 219
E ++++LG ++L+ G + + I A K D I AD H + LS S +
Sbjct: 269 EEELAEWLGYSRALLFISGFAANQAVIAAMMAKEDRIAADRLSHASLLEAASLSPSQLRR 90
Query: 220 FKHNNMDSLRETL 232
F HN++ L L
Sbjct: 89 FAHNDVTHLARLL 51
>BI272653 weakly similar to PIR|E84871|E848 probable polygalacturonase
[imported] - Arabidopsis thaliana, partial (6%)
Length = 345
Score = 30.0 bits (66), Expect = 1.2
Identities = 27/95 (28%), Positives = 44/95 (45%), Gaps = 7/95 (7%)
Frame = +1
Query: 57 YKPPKRPLS-------NKEIDELCDEWVPQPLIPSLNDEMPYEPPVLESAAGPHTIVNGK 109
YKPP+RPLS N I + +W Q ++ L + + PV++SAA G
Sbjct: 1 YKPPRRPLSSPNC*KDNYFITSIEAQWPLQRVLLFLFCSLHW--PVMQSAAPKPRPPVGH 174
Query: 110 EVVNFASANYLGLIGHQKLLDSCSSALEKYGVGSC 144
+V ++ L+ +KL++ S + GV C
Sbjct: 175 DVFGVRKSSKDVLLPGEKLVNVLSFGAKGDGVTDC 279
>BI264649
Length = 678
Score = 27.7 bits (60), Expect = 6.0
Identities = 17/60 (28%), Positives = 30/60 (49%), Gaps = 1/60 (1%)
Frame = +3
Query: 181 TMFSAIPAFSKKGDIIVA-DEGVHWGIQNGLYLSRSTVVYFKHNNMDSLRETLENITSIY 239
T+ S+ P S K + D G+ + + +Y ++ +F +D LRE+L + SIY
Sbjct: 159 TVVSSRPVSSGKSHKLTGVDHGMSYHTLHLIYYYKNEEKWFGSFELDPLRESLSEVLSIY 338
>TC82414 similar to PIR|T49915|T49915 pre-mRNA splicing factor ATP-dependent
RNA helicase-like protein - Arabidopsis thaliana,
partial (13%)
Length = 740
Score = 27.3 bits (59), Expect = 7.9
Identities = 16/45 (35%), Positives = 18/45 (39%), Gaps = 11/45 (24%)
Frame = -3
Query: 61 KRPLSNKEIDELCDEWV-----------PQPLIPSLNDEMPYEPP 94
K P N E+ D W P PL PSLN P+ PP
Sbjct: 474 KNPFLNLELKSAADSWPFFPSPSVYFLSPTPLAPSLNS--PFPPP 346
>TC86934 similar to PIR|T04630|T04630 synaptobrevin homolog F10N7.40 -
Arabidopsis thaliana, complete
Length = 1208
Score = 27.3 bits (59), Expect = 7.9
Identities = 19/61 (31%), Positives = 27/61 (44%), Gaps = 12/61 (19%)
Frame = +2
Query: 232 LENITSIYKR------------TKNLRRYIVIEALYQNSGQIAPLDEIIKLKEKYCFRIL 279
LE++TSIY + L ++ +AL Q SG + + KEKYC L
Sbjct: 821 LEDVTSIYLQLWRELPFVFISIVAGLSTCVLAKALQQISGVLGDIFHYRGFKEKYCVSYL 1000
Query: 280 L 280
L
Sbjct: 1001L 1003
>BQ144513 similar to GP|995935|gb|AA MAZ {Homo sapiens}, partial (3%)
Length = 1447
Score = 27.3 bits (59), Expect = 7.9
Identities = 16/44 (36%), Positives = 22/44 (49%), Gaps = 2/44 (4%)
Frame = -1
Query: 77 VPQPLIP--SLNDEMPYEPPVLESAAGPHTIVNGKEVVNFASAN 118
+P P P SL + +PP + G TIVN + V+ F S N
Sbjct: 676 LPTPAFPPTSLISLLNLQPPGVNQKGGALTIVNRRGVLGFPSRN 545
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.321 0.141 0.422
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 10,405,493
Number of Sequences: 36976
Number of extensions: 158249
Number of successful extensions: 741
Number of sequences better than 10.0: 20
Number of HSP's better than 10.0 without gapping: 736
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 741
length of query: 316
length of database: 9,014,727
effective HSP length: 96
effective length of query: 220
effective length of database: 5,465,031
effective search space: 1202306820
effective search space used: 1202306820
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 58 (26.9 bits)
Medicago: description of AC137824.11


