

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC136953.6 + phase: 0
(268 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
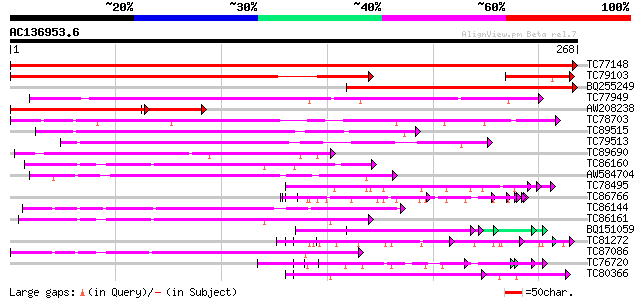
Score E
Sequences producing significant alignments: (bits) Value
TC77148 ENOD20 553 e-158
TC79103 ENOD16 211 3e-55
BQ255249 weakly similar to SP|P93329|NO20 Early nodulin 20 precu... 149 8e-37
TC77949 weakly similar to GP|11762218|gb|AAG40387.1 AT4g27520 {A... 128 3e-30
AW208238 SP|P93328|NO16 Early nodulin 16 precursor (N-16). [Barr... 100 3e-30
TC78703 homologue to GP|13129456|gb|AAK13114.1 Putative retroele... 117 6e-27
TC89515 weakly similar to GP|11994144|dbj|BAB01165. contains sim... 99 2e-21
TC79513 weakly similar to GP|9885806|gb|AAG01535.1| stellacyanin... 96 1e-20
TC89690 weakly similar to PIR|G84643|G84643 similar to early nod... 93 1e-19
TC86160 weakly similar to PIR|T01605|T01605 phytocyanin At2g4479... 89 2e-18
AW584704 similar to SP|P05790|FBOH Fibroin heavy chain precursor... 81 4e-16
TC78495 homologue to PIR|S53504|S53504 extensin-like protein S3 ... 80 6e-16
TC86766 weakly similar to PIR|T06291|T06291 extensin homolog T9E... 80 8e-16
TC86144 weakly similar to GP|10727325|gb|AAF50995.2 CG15635 gene... 80 1e-15
TC86161 weakly similar to PIR|T01605|T01605 phytocyanin At2g4479... 79 2e-15
BQ151059 similar to GP|7291525|gb|A CG9888-PA {Drosophila melano... 77 5e-15
TC81272 similar to PIR|T05225|T05225 extensin homolog F17I5.160 ... 77 9e-15
TC87086 similar to SP|Q41001|BCP_PEA Blue copper protein precurs... 75 2e-14
TC76720 similar to GP|11121502|emb|CAC14888. putative extensin {... 75 3e-14
TC80366 similar to GP|13877617|gb|AAK43886.1 protein kinase-like... 74 5e-14
>TC77148 ENOD20
Length = 1108
Score = 553 bits (1424), Expect = e-158
Identities = 268/268 (100%), Positives = 268/268 (100%)
Frame = +3
Query: 1 MSSSSPILLMFIFSIWMLISYSESTDYLVGDSENSWKFPLPTRHALTRWASNYQFIVGDT 60
MSSSSPILLMFIFSIWMLISYSESTDYLVGDSENSWKFPLPTRHALTRWASNYQFIVGDT
Sbjct: 21 MSSSSPILLMFIFSIWMLISYSESTDYLVGDSENSWKFPLPTRHALTRWASNYQFIVGDT 200
Query: 61 ITFQYNNKTESVHEVEEEDYDRCGIRGEHVDHYDGNTMVVLKKTGIHHFISGKKRHCRLG 120
ITFQYNNKTESVHEVEEEDYDRCGIRGEHVDHYDGNTMVVLKKTGIHHFISGKKRHCRLG
Sbjct: 201 ITFQYNNKTESVHEVEEEDYDRCGIRGEHVDHYDGNTMVVLKKTGIHHFISGKKRHCRLG 380
Query: 121 LKLAVVVMVAPVLSSPPPPPSPPTPRSSTPIPHPPRRSLPSPPSPSPSPSPSPSPSPSPR 180
LKLAVVVMVAPVLSSPPPPPSPPTPRSSTPIPHPPRRSLPSPPSPSPSPSPSPSPSPSPR
Sbjct: 381 LKLAVVVMVAPVLSSPPPPPSPPTPRSSTPIPHPPRRSLPSPPSPSPSPSPSPSPSPSPR 560
Query: 181 STPIPHPRKRSPASPSPSPSLSKSPSPSESPSLAPSPSDSVASLAPSSSPSDESPSPAPS 240
STPIPHPRKRSPASPSPSPSLSKSPSPSESPSLAPSPSDSVASLAPSSSPSDESPSPAPS
Sbjct: 561 STPIPHPRKRSPASPSPSPSLSKSPSPSESPSLAPSPSDSVASLAPSSSPSDESPSPAPS 740
Query: 241 PSSSGSKGGGAGHGFLEVSIAMMMFLIF 268
PSSSGSKGGGAGHGFLEVSIAMMMFLIF
Sbjct: 741 PSSSGSKGGGAGHGFLEVSIAMMMFLIF 824
>TC79103 ENOD16
Length = 829
Score = 211 bits (536), Expect = 3e-55
Identities = 102/172 (59%), Positives = 125/172 (72%)
Frame = +2
Query: 1 MSSSSPILLMFIFSIWMLISYSESTDYLVGDSENSWKFPLPTRHALTRWASNYQFIVGDT 60
M+SSSPILLM IFS+W+LIS+SESTDYL+GDS NSWK PLP+R A RWAS ++F VGDT
Sbjct: 26 MASSSPILLMIIFSMWLLISHSESTDYLIGDSHNSWKVPLPSRRAFARWASAHEFTVGDT 205
Query: 61 ITFQYNNKTESVHEVEEEDYDRCGIRGEHVDHYDGNTMVVLKKTGIHHFISGKKRHCRLG 120
I F+Y+N+TESVHEV E DY C GEHV+H+DGNT VVL K G++HFISG KRHC++G
Sbjct: 206 ILFEYDNETESVHEVNEHDYIMCHTNGEHVEHHDGNTKVVLDKIGVYHFISGTKRHCKMG 385
Query: 121 LKLAVVVMVAPVLSSPPPPPSPPTPRSSTPIPHPPRRSLPSPPSPSPSPSPS 172
LKLAVVV ++ + PP ++P PPSPSPSP+ S
Sbjct: 386 LKLAVVV------------------QNKHDLVLPPLITMPMPPSPSPSPNSS 487
Score = 49.3 bits (116), Expect = 2e-06
Identities = 25/36 (69%), Positives = 29/36 (80%), Gaps = 3/36 (8%)
Frame = +2
Query: 235 PSPAPSPSSSGSKGGGAGHGF---LEVSIAMMMFLI 267
PSP+PSP+SSG+KGG AG GF L VS+ MMMFLI
Sbjct: 458 PSPSPSPNSSGNKGGAAGLGFIMWLGVSLVMMMFLI 565
>BQ255249 weakly similar to SP|P93329|NO20 Early nodulin 20 precursor (N-20).
[Barrel medic] {Medicago truncatula}, partial (40%)
Length = 609
Score = 149 bits (377), Expect = 8e-37
Identities = 75/109 (68%), Positives = 75/109 (68%)
Frame = +1
Query: 160 PSPPSPSPSPSPSPSPSPSPRSTPIPHPRKRSPASPSPSPSLSKSPSPSESPSLAPSPSD 219
P PP P P P P P PSP PIPHP P PSPSPSL K PSPSESPSL PSPSD
Sbjct: 1 PXPPLXXPLPLPLPLPLPSPWXNPIPHPWXXXPGLPSPSPSLXKXPSPSESPSLXPSPSD 180
Query: 220 SVASLAPSSSPSDESPSPAPSPSSSGSKGGGAGHGFLEVSIAMMMFLIF 268
SV SL P SPS E P P PSPS GSKGGGAGHGFLEVSIA FLIF
Sbjct: 181 SVGSLGPLFSPSKELPLPGPSPSLFGSKGGGAGHGFLEVSIAKKKFLIF 327
>TC77949 weakly similar to GP|11762218|gb|AAG40387.1 AT4g27520 {Arabidopsis
thaliana}, partial (34%)
Length = 1297
Score = 128 bits (321), Expect = 3e-30
Identities = 82/252 (32%), Positives = 127/252 (49%), Gaps = 9/252 (3%)
Frame = +3
Query: 10 MFIFSIWMLISYSESTDYLVGDSENSWKFPLPTRHALTRWASNYQFIVGDTITFQYNNKT 69
+F +++S S++ + VG + + L +WA +F + DTI F+Y +
Sbjct: 138 LFFVLFTLVVSTSQAFKFFVGGKDG---WTLNPSENYNQWAGRNRFQISDTIVFKYKKGS 308
Query: 70 ESVHEVEEEDYDRCGIRGEHVDHYDGNTMVVLKKTGIHHFISGKKRHCRLGLKLAVVVMV 129
+SV EV++EDY++C DG T L + G +FISGK ++C G KL +VV+
Sbjct: 309 DSVLEVKKEDYEKCNKTNPIKKFEDGETEFTLDRAGPFYFISGKDQNCENGQKLTLVVIS 488
Query: 130 APVLSSPPPPP----SPPTPRSSTPIPHPPRRSLPSPPS--PSPSPSPSPSPSPSPRSTP 183
S P P SPP+P +T P P S PSP + P+ SP P+ P+ S S
Sbjct: 489 PRTPKSSPSPSAGGLSPPSPSPTTTTP-SPSGSPPSPVAIPPASSPVPTSGPTASSPSPV 665
Query: 184 IPHPRKRSPASPSPSPSLSKSPSPSESPSLAPSPSDSVASLAPSSSPSDES---PSPAPS 240
+ P P + SPSP +S P+ S +PSP+ +L+P+ PS + P+P P
Sbjct: 666 VSTPPAGGPMASSPSPVVSTPPAGGPMAS-SPSPAGGPPALSPAGGPSTAAGGPPAPGPG 842
Query: 241 PSSSGSKGGGAG 252
+++ GGAG
Sbjct: 843 GAATSPGPGGAG 878
>AW208238 SP|P93328|NO16 Early nodulin 16 precursor (N-16). [Barrel medic]
{Medicago truncatula}, partial (51%)
Length = 402
Score = 100 bits (250), Expect(2) = 3e-30
Identities = 46/66 (69%), Positives = 55/66 (82%)
Frame = +3
Query: 1 MSSSSPILLMFIFSIWMLISYSESTDYLVGDSENSWKFPLPTRHALTRWASNYQFIVGDT 60
M+SSSPILLM IFS+W+LIS+SESTDYL+GDS NSWK PLP+R A RWAS ++F VGDT
Sbjct: 27 MASSSPILLMIIFSMWLLISHSESTDYLIGDSHNSWKVPLPSRRAFARWASAHEFTVGDT 206
Query: 61 ITFQYN 66
I Y+
Sbjct: 207IRKPYS 224
Score = 48.1 bits (113), Expect(2) = 3e-30
Identities = 19/31 (61%), Positives = 24/31 (77%)
Frame = +2
Query: 63 FQYNNKTESVHEVEEEDYDRCGIRGEHVDHY 93
F+Y+N+TESVHEV E DY C GEHV+H+
Sbjct: 308 FEYDNETESVHEVNEHDYIMCHTNGEHVEHH 400
>TC78703 homologue to GP|13129456|gb|AAK13114.1 Putative retroelement pol
polyprotein {Oryza sativa} [Oryza sativa (japonica
cultivar-group)], partial (2%)
Length = 1235
Score = 117 bits (292), Expect = 6e-27
Identities = 89/280 (31%), Positives = 134/280 (47%), Gaps = 20/280 (7%)
Frame = +3
Query: 1 MSSSSPILLMFIFSIWMLISYSESTDYLVGDSENSWKFPL-PTRHALTRWASNYQFIVGD 59
M++ L++F + +L+ ++ +++VG + W P P + +WA +F VGD
Sbjct: 75 MATIVQALVLFCLLV-LLMHKGDAYEFVVG-GQKGWSAPSDPNANPYNQWAEKSRFQVGD 248
Query: 60 TITFQYNNKTESVHEV-EEEDYDRCGIRGEHVDHYDGNTMVVLKKTGIHHFISGKKRHCR 118
++ F Y + +SV +V ++DY+ C DG+T++ L K+G H+FISG K +C
Sbjct: 249 SLVFNYQSGQDSVIQVTSQQDYENCNTDASSEKSSDGHTVIKLIKSGPHYFISGNKNNCL 428
Query: 119 LGLKLAVVVMVAPVLSSPPPPPSPPTPRSSTPIPHPPRRSLPSPPSPSPSPSPSPSPSPS 178
KL V+V+ + T ++S SPPSPSPS +PSPSP S
Sbjct: 429 QNEKLLVIVL------------ADRTNKNSN--------QTTSPPSPSPSVAPSPSPLSS 548
Query: 179 PRS---TPIPHPRKRSPASPSPSPSLSKS---PSPSESPSLAPSPSDSVAS--------- 223
S TPIP P + +S PSP L S PSP + +L P P V S
Sbjct: 549 HSSDALTPIPPPSPLNGSSTPPSPVLDGSFPPPSPLDGSTLTPPPVQQVGSSPPPLGTDV 728
Query: 224 ---LAPSSSPSDESPSPAPSPSSSGSKGGGAGHGFLEVSI 260
+ P+ SP E P P + +SS G G L VS+
Sbjct: 729 TNPITPTQSPVSEPPPP--NAASSILVSFGCSVGALMVSL 842
>TC89515 weakly similar to GP|11994144|dbj|BAB01165. contains similarity to
nodulin~gene_id:K10D20.11 {Arabidopsis thaliana},
partial (43%)
Length = 854
Score = 98.6 bits (244), Expect = 2e-21
Identities = 61/189 (32%), Positives = 92/189 (48%), Gaps = 7/189 (3%)
Frame = +2
Query: 13 FSIWMLISYSESTDYLVGDSENSWKFPLPTRHALTRWASNYQFIVGDTITFQYNNKTESV 72
F + ++I S + D++VG + W P + + +WA +F VGD++ F Y + +SV
Sbjct: 95 FCLMLMIHKSAAYDFIVG-GQKGWSVPSDSNNPFNQWAEKSRFQVGDSLVFNYQSGKDSV 271
Query: 73 HEVEEEDYDRCGIRGEHVDHYDGNTMVVLKKTGIHHFISGKKRHCRLGLKLAVVVMVAPV 132
V+ EDY C DG+T+ L ++G H FISG K +C K+ V+V+
Sbjct: 272 LYVKSEDYASCNTGSPITKFSDGHTVFKLNQSGPHFFISGNKDNCLKNEKVTVIVLSDRS 451
Query: 133 LSSPPPPPSPPTPRSSTPIPHPPRRSLPSPPSPSPSPSPSPSPSPSPRSTPIP------- 185
++ S T ++S P P+ S SPPSP+PS SP P TP P
Sbjct: 452 NNN----NSSNTNQTSNATPPSPQSS--SPPSPAPSNQEGQSPPPDTNQTPPPTATSDHD 613
Query: 186 HPRKRSPAS 194
HP + AS
Sbjct: 614 HPPRNGAAS 640
>TC79513 weakly similar to GP|9885806|gb|AAG01535.1| stellacyanin-like
protein CASLP1 precursor {Capsicum annuum}, partial
(33%)
Length = 1093
Score = 95.9 bits (237), Expect = 1e-20
Identities = 63/207 (30%), Positives = 96/207 (45%), Gaps = 3/207 (1%)
Frame = +3
Query: 25 TDYLVGDSENSWKFPLPTRHALTRWASNYQFIVGDTITFQYNNKTESVHEVEEEDYDRCG 84
T ++VGD+ W P T WASN F VGDT+ F Y + V +V + YD C
Sbjct: 471 TRHVVGDT-TGWTIPTNGASFYTNWASNKTFTVGDTLVFNYASGQHDVAKVTKTAYDSCN 647
Query: 85 IRGEHVDHYDGNTMVVLKKTGIHHFISGKKRHCRLGLKLAVVVMVAPVLSSPPPPPSPPT 144
+ V L +TG +FI HC G KL++ V+ A P S PT
Sbjct: 648 GANTLFTLTNSPATVTLNETGQQNFICAVPGHCSAGQKLSINVVKASA-----SPVSAPT 812
Query: 145 PRSSTPIPHPPRRSLPSPPSPSPSPSPSPSPSPSPRSTPIPHPRKRSPASPSPSPSLSKS 204
P +S PP +P+P+P P+ SP+P A+P+P+PS + S
Sbjct: 813 PSAS-------------PPKATPAPTPVPAKSPAPTKA----------ATPAPAPSTTAS 923
Query: 205 PSPSESPS---LAPSPSDSVASLAPSS 228
P+P+ +P+ + + D++ + PS+
Sbjct: 924 PTPAPAPATGRVTYTVGDTIGWIIPSN 1004
>TC89690 weakly similar to PIR|G84643|G84643 similar to early nodulins
[imported] - Arabidopsis thaliana, partial (56%)
Length = 732
Score = 92.8 bits (229), Expect = 1e-19
Identities = 59/160 (36%), Positives = 85/160 (52%), Gaps = 8/160 (5%)
Frame = +3
Query: 3 SSSPILLMFIFSIWMLISYSESTDYLVGDSENSWKFPLPTRHALTRWASNYQFIVGDTIT 62
SS +L++F+ + + D L+G ++WK P +L +WAS+ +F VGD +
Sbjct: 15 SSCSLLVLFV----LFGCAFAAKDILLGGKTDAWKVPSSESDSLNKWASSVRFQVGDHLI 182
Query: 63 FQYNNKTESVHEVEEEDYDRCGIRGEHVDHY-DGNTMVVLKKTGIHHFISGKKRHCRLGL 121
+Y +SV +V +EDYD C I + + HY DGNT V +G +++ISG+K HC G
Sbjct: 183 LKYEAGKDSVLQVSKEDYDSCNI-SKPIKHYNDGNTKVRFDHSGPYYYISGEKGHCEKGQ 359
Query: 122 KLAVVVMVAPVLSSP----PPPPSPPT---PRSSTPIPHP 154
KL VVVM S P P PSP P +S P P
Sbjct: 360 KLTVVVMSLKGGSRPIVAFSPSPSPAEVEGPAASAVAPAP 479
Score = 27.7 bits (60), Expect = 4.8
Identities = 18/56 (32%), Positives = 31/56 (55%)
Frame = +3
Query: 211 PSLAPSPSDSVASLAPSSSPSDESPSPAPSPSSSGSKGGGAGHGFLEVSIAMMMFL 266
P +A SPS S A + P+ + +PAP+ ++ +GGG F+ V + + M+L
Sbjct: 402 PIVAFSPSPSPAEV---EGPAASAVAPAPTSGAAVLQGGGV---FVAVGVFVAMWL 551
>TC86160 weakly similar to PIR|T01605|T01605 phytocyanin At2g44790
[imported] - Arabidopsis thaliana, partial (34%)
Length = 918
Score = 88.6 bits (218), Expect = 2e-18
Identities = 60/172 (34%), Positives = 86/172 (49%), Gaps = 6/172 (3%)
Frame = +3
Query: 8 LLMFIFSIWMLISYSESTDYLVGDSENSWKFPLPTRHALTRWASNYQFIVGDTITFQYNN 67
++ F + + Y+ +TD+ VGD+ N W + T+WAS F VGD + F+Y +
Sbjct: 132 MIASFFVLLLAFPYAFATDFTVGDA-NGWNLGVD----YTKWASGKTFKVGDNLVFKYGS 296
Query: 68 KTESVHEVEEEDYDRCGIRGEHVDHYDGNTMVVLKKTGIHHFISGKKRHCRL--GLKLAV 125
+ V EV+E DY C ++ GN+ V L K G +FI HC G+KL V
Sbjct: 297 -SHQVDEVDESDYKSCTSSNAIKNYAGGNSKVPLTKAGKIYFICPTLGHCTSTGGMKLEV 473
Query: 126 VVMVAPVL----SSPPPPPSPPTPRSSTPIPHPPRRSLPSPPSPSPSPSPSP 173
V+ A +PPP SP T S+TP P + PS PS + S +PSP
Sbjct: 474 NVVAASTTPTPSGTPPPTKSPSTTPSTTPSTTP--STTPSAPSETNSTTPSP 623
Score = 37.7 bits (86), Expect = 0.005
Identities = 21/64 (32%), Positives = 33/64 (50%)
Frame = +3
Query: 193 ASPSPSPSLSKSPSPSESPSLAPSPSDSVASLAPSSSPSDESPSPAPSPSSSGSKGGGAG 252
AS +P+PS +P P++SPS PS + S S+PS+ + + P +G+ G G
Sbjct: 486 ASTTPTPS--GTPPPTKSPSTTPSTTPSTTPSTTPSAPSETNSTTPSPPKDNGAVGVSNG 659
Query: 253 HGFL 256
L
Sbjct: 660 VSLL 671
>AW584704 similar to SP|P05790|FBOH Fibroin heavy chain precursor (Fib-H)
(H-fibroin). [Silk moth] {Bombyx mori}, partial (0%)
Length = 679
Score = 81.3 bits (199), Expect = 4e-16
Identities = 58/188 (30%), Positives = 90/188 (47%), Gaps = 14/188 (7%)
Frame = +1
Query: 10 MFIFSIWMLI--SYSESTDYLVGDSENSWKFPLPTRHALTRWASNYQFIVGDTITFQYNN 67
+F+F++ I + + + D++VGD E+ W + + WA+N F +GDT+TF+Y
Sbjct: 70 LFLFALIATIFSTMAVAKDFVVGD-ESGWTLGVDYQ----AWAANKVFRLGDTLTFKYVA 234
Query: 68 KTESVHEVEEEDYDRCGIRGEHVDHYDGNTMVVLKKTGIHHFISGKKRHCRLGLKLAVVV 127
++V V D+ C + G+ + L G +ISG HC G KL + V
Sbjct: 235 WKDNVVRVNGSDFQSCSVPWAAPVLTSGHDKIALTTYGRRWYISGVANHCENGQKLFINV 414
Query: 128 MVAPVLSSPPPPPSPPTPRSSTPIPHPPRRSLPSPPSPSP------------SPSPSPSP 175
+ P P PS P S++P P P +PPS +P SP PSPSP
Sbjct: 415 L--PKQDGWYPAPSSP---SASPSPSPVPAPEAAPPSNAPWAASVQTSELTWSPVPSPSP 579
Query: 176 SPSPRSTP 183
+P+P + P
Sbjct: 580 TPAPEAAP 603
>TC78495 homologue to PIR|S53504|S53504 extensin-like protein S3 - alfalfa,
partial (97%)
Length = 1049
Score = 80.5 bits (197), Expect = 6e-16
Identities = 52/132 (39%), Positives = 69/132 (51%), Gaps = 4/132 (3%)
Frame = +3
Query: 131 PVLSSPPPPPSPPTPRSSTPIPHPPRRSLPSPPSPSP-SPSPSPSPSPSPRSTPIPHPRK 189
PV SPPP P P P STP P P + P P SP P +P P+ P +P P P
Sbjct: 432 PVQQSPPPTPLTPPPVQSTPPPASPPPASPPPFSPPPATPPPATPPPATPPPALTPTPLS 611
Query: 190 RSPA-SPSPSPSLSKSPSPSESPSLAPS--PSDSVASLAPSSSPSDESPSPAPSPSSSGS 246
PA +P+P+P+ KS +P+ +P L+PS P+ ++SL+PS SPS S A S
Sbjct: 612 SPPATTPAPAPAKLKSKAPALAPVLSPSDAPAPGLSSLSPSISPSGTDDSGAEKLWSHKM 791
Query: 247 KGGGAGHGFLEV 258
G G FL +
Sbjct: 792 VGLVFGCAFLSL 827
Score = 77.8 bits (190), Expect = 4e-15
Identities = 47/120 (39%), Positives = 58/120 (48%), Gaps = 3/120 (2%)
Frame = +3
Query: 131 PVLSSPPPPPSPPTPRSSTPIPHPPRRSLPSPPSPSPSP---SPSPSPSPSPRSTPIPHP 187
PV SSPPP S P P S+P PP +S P P SP P SP P+P P P P
Sbjct: 327 PVQSSPPPVQSSPPPLQSSP---PPAQSTPPPVQSSPPPVQQSPPPTPLTPPPVQSTPPP 497
Query: 188 RKRSPASPSPSPSLSKSPSPSESPSLAPSPSDSVASLAPSSSPSDESPSPAPSPSSSGSK 247
PASP P SP P+ P P P+ +L P+ S + +PAP+P+ SK
Sbjct: 498 ASPPPASPPPF-----SPPPATPPPATPPPATPPPALTPTPLSSPPATTPAPAPAKLKSK 662
Score = 76.6 bits (187), Expect = 9e-15
Identities = 55/134 (41%), Positives = 71/134 (52%), Gaps = 13/134 (9%)
Frame = +3
Query: 131 PVLSSPPPPPSPPTPRSSTPIP---HPPRRSLPSPPSPSPSPSPSPSP-SPSPRSTPIPH 186
P+ SSPPP S P P S+P P PP L PP S P SP P SP P S P
Sbjct: 369 PLQSSPPPAQSTPPPVQSSPPPVQQSPPPTPLTPPPVQSTPPPASPPPASPPPFSPPPAT 548
Query: 187 PRKRSPASPSPSPSLSKSP---SPSESPSLAPSPSDSVA-SLAPSSSPSDESPSP----- 237
P +P +P P+L+ +P P+ +P+ AP+ S A +LAP SPSD +P+P
Sbjct: 549 PPPATPPPATPPPALTPTPLSSPPATTPAPAPAKLKSKAPALAPVLSPSD-APAPGLSSL 725
Query: 238 APSPSSSGSKGGGA 251
+PS S SG+ GA
Sbjct: 726 SPSISPSGTDDSGA 767
>TC86766 weakly similar to PIR|T06291|T06291 extensin homolog T9E8.80 -
Arabidopsis thaliana, partial (18%)
Length = 1260
Score = 80.1 bits (196), Expect = 8e-16
Identities = 54/118 (45%), Positives = 57/118 (47%), Gaps = 9/118 (7%)
Frame = +2
Query: 129 VAPVLSSPPPPP--SPPTPRSSTPIPHPPRRSLPSPPSPSPSPSPSPSPSPSPRSTPIPH 186
V P LS PPPPP SPP P S P P PP P PP P P P P SP+P TP
Sbjct: 200 VYPYLSPPPPPPVHSPPPPVYSPPPPSPPPCVEPPPPPPPPCVEPPPPSSPAPHQTPYHP 379
Query: 187 PRKRSP------ASPSPSPSLSKSPSPSESPSLA-PSPSDSVASLAPSSSPSDESPSP 237
P SP A PSP P + SP P SP A PSP V S +P P E P P
Sbjct: 380 PPSPSPPPSPVYAYPSPPPPVYTSPPP--SPVYAYPSPPPPVYS-SPPPPPVYEGPIP 544
Score = 75.9 bits (185), Expect = 2e-14
Identities = 49/117 (41%), Positives = 58/117 (48%), Gaps = 4/117 (3%)
Frame = +2
Query: 130 APVLSSPPPPP----SPPTPRSSTPIPHPPRRSLPSPPSPSPSPSPSPSPSPSPRSTPIP 185
APV S PPP P SPP P + +P P PP P P SP+P P SP P P P
Sbjct: 74 APVFSPPPPVPYYYSSPPPPPAHSP-PPPPXSPPPPPHSPTPPVYPYLSPPPPP-PVHSP 247
Query: 186 HPRKRSPASPSPSPSLSKSPSPSESPSLAPSPSDSVASLAPSSSPSDESPSPAPSPS 242
P SP PSP P + + P P P + P P +S AP +P PSP+P PS
Sbjct: 248 PPPVYSPPPPSPPPCV-EPPPPPPPPCVEPPPP---SSPAPHQTPYHPPPSPSPPPS 406
Score = 73.2 bits (178), Expect = 1e-13
Identities = 54/132 (40%), Positives = 59/132 (43%), Gaps = 17/132 (12%)
Frame = +2
Query: 131 PVLSSPPPPPSPPTPRSS---TPIPHPPRRSLP----SPPSPSPSPSPSPSP-------S 176
P SPPPPP PTP +P P PP S P SPP PSP P P P
Sbjct: 152 PPPXSPPPPPHSPTPPVYPYLSPPPPPPVHSPPPPVYSPPPPSPPPCVEPPPPPPPPCVE 331
Query: 177 PSPRSTPIPHPRKRSPASPSPSPSLSKSP---SPSESPSLAPSPSDSVASLAPSSSPSDE 233
P P S+P PH ++P P PSPS SP PS P + SP S PS P
Sbjct: 332 PPPPSSPAPH---QTPYHPPPSPSPPPSPVYAYPSPPPPVYTSPPPSPVYAYPSPPPPVY 502
Query: 234 SPSPAPSPSSSG 245
S SP P P G
Sbjct: 503 S-SPPPPPVYEG 535
Score = 72.0 bits (175), Expect = 2e-13
Identities = 49/124 (39%), Positives = 56/124 (44%), Gaps = 10/124 (8%)
Frame = +2
Query: 131 PVLSSPPPPPSPPTPRSSTPIPHPPRRSLPSPPSPSPSPSPSPSPSPSPRS-TPIPHPRK 189
P +SPPPPP PP P S P P P S P PP P P SP P P S TP +P
Sbjct: 38 PPPNSPPPPP-PPAPVFSPPPPVPYYYSSPPPPPAHSPPPPPXSPPPPPHSPTPPVYPYL 214
Query: 190 RSPASP---SPSPSLSKSPSPSESPSLAPSPSDSVASLAPS--SSPSDES----PSPAPS 240
P P SP P + P PS P + P P + P SSP+ P P+PS
Sbjct: 215 SPPPPPPVHSPPPPVYSPPPPSPPPCVEPPPPPPPPCVEPPPPSSPAPHQTPYHPPPSPS 394
Query: 241 PSSS 244
P S
Sbjct: 395 PPPS 406
Score = 61.2 bits (147), Expect = 4e-10
Identities = 44/110 (40%), Positives = 50/110 (45%), Gaps = 5/110 (4%)
Frame = +2
Query: 137 PPPP----SPPTPRSSTPIPHPPRRSLPSPPSPSPSPSPSPSPSPSPRSTPIPHPRKRSP 192
P PP SPP P ++P P PP + SPP P P SP P P+ S P P P
Sbjct: 2 PTPPYCVRSPPPPPPNSPPPPPPPAPVFSPPPPVPYYYSSPPPPPA-HSPPPP------P 160
Query: 193 ASPSPSPSLSKSPSPSESPSLAPSPSDSVASLAPS-SSPSDESPSPAPSP 241
SP P P SP+P P L+P P V S P SP SP P P
Sbjct: 161 XSPPPPP---HSPTPPVYPYLSPPPPPPVHSPPPPVYSPPPPSPPPCVEP 301
Score = 60.5 bits (145), Expect = 7e-10
Identities = 40/104 (38%), Positives = 49/104 (46%), Gaps = 4/104 (3%)
Frame = +2
Query: 131 PVLSSPPPPPSPPT---PRSSTPIPHPPRRSLPSPPSPSPSPSPS-PSPSPSPRSTPIPH 186
P PPPPP PP P S+P PH P PSP PSP + PSP P ++P P
Sbjct: 284 PPCVEPPPPPPPPCVEPPPPSSPAPHQTPYHPPPSPSPPPSPVYAYPSPPPPVYTSPPPS 463
Query: 187 PRKRSPASPSPSPSLSKSPSPSESPSLAPSPSDSVASLAPSSSP 230
P A PSP P + SP P P + P V ++ +S P
Sbjct: 464 P---VYAYPSPPPPVYSSPPP---PPVYEGPIPPVFGISYASPP 577
Score = 41.2 bits (95), Expect = 4e-04
Identities = 32/74 (43%), Positives = 35/74 (47%), Gaps = 5/74 (6%)
Frame = +2
Query: 131 PVLSSPPPP-----PSPPTPRSSTPIPHPPRRSLPSPPSPSPSPSPSPSPSPSPRSTPIP 185
P SPPP PSPP P ++P P P + PSPP P S SP P P PIP
Sbjct: 380 PPSPSPPPSPVYAYPSPPPPVYTSP-PPSPVYAYPSPPPPVYS---SPPPPPVYEG-PIP 544
Query: 186 HPRKRSPASPSPSP 199
S ASP P P
Sbjct: 545 PVFGISYASPPPPP 586
>TC86144 weakly similar to GP|10727325|gb|AAF50995.2 CG15635 gene product
{Drosophila melanogaster}, partial (4%)
Length = 922
Score = 79.7 bits (195), Expect = 1e-15
Identities = 61/181 (33%), Positives = 87/181 (47%)
Frame = +3
Query: 7 ILLMFIFSIWMLISYSESTDYLVGDSENSWKFPLPTRHALTRWASNYQFIVGDTITFQYN 66
IL + F+ +L S +E+ D+ VG + W P + WA++ F D + F +
Sbjct: 114 ILFVVAFAAAILES-TEAADHTVGGT-TGWSVPSGASF-YSDWAASNTFKQNDVLVFNFA 284
Query: 67 NKTESVHEVEEEDYDRCGIRGEHVDHYDGNTMVVLKKTGIHHFISGKKRHCRLGLKLAVV 126
+V EV + D+D C I + G V L +TG +FI + HC G KL+V
Sbjct: 285 GG-HTVAEVSKADFDNCNINQNGLVITTGPARVTLNRTGDFYFICTIQGHCSSGQKLSV- 458
Query: 127 VMVAPVLSSPPPPPSPPTPRSSTPIPHPPRRSLPSPPSPSPSPSPSPSPSPSPRSTPIPH 186
V +S P PPS PTP +STP P S +P P+ +PSPS P +TP P
Sbjct: 459 ----KVSASTPSPPS-PTPPTSTPPTSTPPTSGTTPTPPTNGGTPSPSSPTGPDATP-PS 620
Query: 187 P 187
P
Sbjct: 621 P 623
Score = 31.6 bits (70), Expect = 0.34
Identities = 15/55 (27%), Positives = 30/55 (54%), Gaps = 1/55 (1%)
Frame = +3
Query: 191 SPASPSPSPSLSKSPSPSESPS-LAPSPSDSVASLAPSSSPSDESPSPAPSPSSS 244
+P+ PSP+P S P+ + S P+P + + +PSS ++ P+P +++
Sbjct: 474 TPSPPSPTPPTSTPPTSTPPTSGTTPTPPTNGGTPSPSSPTGPDATPPSPGSATT 638
Score = 26.9 bits (58), Expect = 8.3
Identities = 20/64 (31%), Positives = 30/64 (46%), Gaps = 6/64 (9%)
Frame = +3
Query: 189 KRSPASPSPSPSLSKSPSPSESPSLAPSPSDSVASLAPS-----SSPSDESPS-PAPSPS 242
K S ++PSP PSP+ S P+ + + P+ +PS SP+ P +P
Sbjct: 459 KVSASTPSP-------PSPTPPTSTPPTSTPPTSGTTPTPPTNGGTPSPSSPTGPDATPP 617
Query: 243 SSGS 246
S GS
Sbjct: 618 SPGS 629
>TC86161 weakly similar to PIR|T01605|T01605 phytocyanin At2g44790
[imported] - Arabidopsis thaliana, partial (17%)
Length = 736
Score = 78.6 bits (192), Expect = 2e-15
Identities = 55/171 (32%), Positives = 81/171 (47%), Gaps = 3/171 (1%)
Frame = +1
Query: 5 SPILLMFIFSIWMLISYSESTDYLVGDSENSWKFPLPTRHALTRWASNYQFIVGDTITFQ 64
S ++ F + + Y+ +TD+ VGD+ N W + T+WAS F VGD + F+
Sbjct: 103 STSMIASFFVLLLAFPYAFATDFTVGDA-NGWTQGVD----YTKWASGKTFKVGDNLVFK 267
Query: 65 YNNKTESVHEVEEEDYDRCGIRGEHVDHYDGNTMVVLKKTGIHHFISGKKRHCRL--GLK 122
Y + V+EV+E Y C + DG++ V L K G +FI HC G+K
Sbjct: 268 YGS-FHQVNEVDESGYKSCSTSNTIKSYDDGDSKVPLTKAGKIYFICPTPGHCTSTGGMK 444
Query: 123 LAVVVMVAPVLSSPPPPPSPPTPRSSTP-IPHPPRRSLPSPPSPSPSPSPS 172
L V V+ A +P P P S+TP P + PSPP + + S S
Sbjct: 445 LEVNVVAASTTPTPSGTPPPTKSPSTTPSAPSETNSTTPSPPKDNGAFSVS 597
Score = 28.9 bits (63), Expect = 2.2
Identities = 15/35 (42%), Positives = 20/35 (56%)
Frame = +1
Query: 193 ASPSPSPSLSKSPSPSESPSLAPSPSDSVASLAPS 227
AS +P+PS +P P++SPS PS S PS
Sbjct: 466 ASTTPTPS--GTPPPTKSPSTTPSAPSETNSTTPS 564
>BQ151059 similar to GP|7291525|gb|A CG9888-PA {Drosophila melanogaster},
partial (26%)
Length = 308
Score = 77.4 bits (189), Expect = 5e-15
Identities = 43/114 (37%), Positives = 43/114 (37%)
Frame = -1
Query: 136 PPPPPSPPTPRSSTPIPHPPRRSLPSPPSPSPSPSPSPSPSPSPRSTPIPHPRKRSPASP 195
PPPPP PP P P P PP P PP P P P P P P P P P P P P P
Sbjct: 308 PPPPPPPPPPPPPPPPPPPP----PPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPP 141
Query: 196 SPSPSLSKSPSPSESPSLAPSPSDSVASLAPSSSPSDESPSPAPSPSSSGSKGG 249
P P P P P P P P P P P P KGG
Sbjct: 140 PPPPPPPPPPPPPPPPPPPPPP-----------PPPPPPPPPPPPPRGFPRKGG 12
Score = 76.6 bits (187), Expect = 9e-15
Identities = 36/85 (42%), Positives = 37/85 (43%)
Frame = -2
Query: 136 PPPPPSPPTPRSSTPIPHPPRRSLPSPPSPSPSPSPSPSPSPSPRSTPIPHPRKRSPASP 195
PPPPP PP P P P PP P PP P P P P P P P P P P P P P
Sbjct: 262 PPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPP 83
Query: 196 SPSPSLSKSPSPSESPSLAPSPSDS 220
P P P P P + P DS
Sbjct: 82 PPPPPPPPPPPPPPPPGVFPGKGDS 8
Score = 75.1 bits (183), Expect = 3e-14
Identities = 37/96 (38%), Positives = 38/96 (39%)
Frame = -2
Query: 136 PPPPPSPPTPRSSTPIPHPPRRSLPSPPSPSPSPSPSPSPSPSPRSTPIPHPRKRSPASP 195
PPPPP PP P P P PP P PP P P P P P P P P P P P P P
Sbjct: 295 PPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPP 116
Query: 196 SPSPSLSKSPSPSESPSLAPSPSDSVASLAPSSSPS 231
P P P P P P P + P S
Sbjct: 115 PPPPPPPPPPPPPPPPPPPPPPPPPPPGVFPGKGDS 8
Score = 73.2 bits (178), Expect = 1e-13
Identities = 36/89 (40%), Positives = 37/89 (41%)
Frame = -3
Query: 136 PPPPPSPPTPRSSTPIPHPPRRSLPSPPSPSPSPSPSPSPSPSPRSTPIPHPRKRSPASP 195
PPPPP PP P P P PP P PP P P P P P P P P P P P P P
Sbjct: 267 PPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPP 88
Query: 196 SPSPSLSKSPSPSESPSLAPSPSDSVASL 224
P P P P P SP + L
Sbjct: 87 PPPPPPPPPPPPPPPPPPGFSPERGIVVL 1
Score = 58.5 bits (140), Expect = 3e-09
Identities = 32/95 (33%), Positives = 32/95 (33%)
Frame = -2
Query: 160 PSPPSPSPSPSPSPSPSPSPRSTPIPHPRKRSPASPSPSPSLSKSPSPSESPSLAPSPSD 219
P PP P P P P P P P P P P P P P P P P P P P P
Sbjct: 298 PPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPP 119
Query: 220 SVASLAPSSSPSDESPSPAPSPSSSGSKGGGAGHG 254
P P P P P P G G G
Sbjct: 118 PPPPPPPPPPPPPPPPPPPPPPPPPPPPGVFPGKG 14
>TC81272 similar to PIR|T05225|T05225 extensin homolog F17I5.160 -
Arabidopsis thaliana, partial (3%)
Length = 757
Score = 76.6 bits (187), Expect = 9e-15
Identities = 53/148 (35%), Positives = 69/148 (45%), Gaps = 11/148 (7%)
Frame = +3
Query: 131 PVLSSPPPPPSP-PTPRSSTPIPHPPRRSLPSPP-----SPSPSPSPSPSPSPSPRSTPI 184
PV P P P+P PT P P P P P +P P P S +P P P ++
Sbjct: 87 PVTVGPVPLPTPIPTGPGGVPQPGPGASGAPQPGPGASGAPQPGPGASGAPQPGPGASGA 266
Query: 185 PHPRKRSPASPSPSPSLSKSPSPSESPSLAPSPSDSVASLAPSSSP-SDESPSPAPSPSS 243
P P + +P P P S +P P S AP P AS AP +P + +P P PSP+
Sbjct: 267 PQPGPGASGAPQPGPDASGAPQPGPGASGAPQPGPG-ASGAPQPAPEASGAPHPGPSPTH 443
Query: 244 SGSKGGG---AGHGFLEVSI-AMMMFLI 267
+ G G A G L VSI A ++FL+
Sbjct: 444 VTTAGSGKSVASFGALAVSIFAGVVFLL 527
Score = 58.9 bits (141), Expect = 2e-09
Identities = 57/173 (32%), Positives = 70/173 (39%), Gaps = 48/173 (27%)
Frame = +1
Query: 135 SPPPPPSPPTP---RSSTPIPH-----------------------PPRRSLPSPPSPSPS 168
SP PP SPP P S P+P P +LP PSP P
Sbjct: 106 SPCPPRSPPAPVVFPSPAPVPAVLLSLALVLPVLLSPALVLPVLPSPVLALPVLPSPVPV 285
Query: 169 PSPSPSPS---PSPRSTPIPHPRKRSPA-------SPSPSPSLSKSPSPSESPSLAPSPS 218
P PSP+ +P S + P SPA SP P P+ P P+ S P+P
Sbjct: 286 PLALPSPALMPVAPLSPALVLPAPLSPALVLPVLPSPPPRPAALLIPDPAPPTSPPPAP- 462
Query: 219 DSVASLAPSSSPSDESPSPAPSPSSSGSKGG------------GAGHGFLEVS 259
SL+P+S+P + SPA SSS SKGG GHG + VS
Sbjct: 463 ---VSLSPASAPLPSASSPA---SSSFSKGGCKSRGRFPFLFSRLGHGLVNVS 603
Score = 55.1 bits (131), Expect = 3e-08
Identities = 37/95 (38%), Positives = 45/95 (46%), Gaps = 11/95 (11%)
Frame = +1
Query: 127 VMVAPVLSSPPPPP-----------SPPTPRSSTPIPHPPRRSLPSPPSPSPSPSPSPSP 175
V+ PVL SP P P +P +P P P P LP PSP P P+ P
Sbjct: 247 VLALPVLPSPVPVPLALPSPALMPVAPLSPALVLPAPLSPALVLPVLPSPPPRPAALLIP 426
Query: 176 SPSPRSTPIPHPRKRSPASPSPSPSLSKSPSPSES 210
P+P ++P P P SPAS +P PS S S S S
Sbjct: 427 DPAPPTSPPPAPVSLSPAS-APLPSASSPASSSFS 528
Score = 47.8 bits (112), Expect = 5e-06
Identities = 42/118 (35%), Positives = 54/118 (45%), Gaps = 20/118 (16%)
Frame = +1
Query: 146 RSSTPIPHPPRRSLPSPPSPSPSPSPSPSPSPSP-----RSTPIPHPRKRSPA-----SP 195
R + P P P +S PSP P P+P PSP+P S + P SPA P
Sbjct: 61 RRTAPSPSLPSQSAPSPCPPRSPPAPVVFPSPAPVPAVLLSLALVLPVLLSPALVLPVLP 240
Query: 196 SPSPSLSKSPSPSESPSLAPSPS-DSVASLAPS------SSPS---DESPSPAPSPSS 243
SP +L PSP P PSP+ VA L+P+ SP+ PSP P P++
Sbjct: 241 SPVLALPVLPSPVPVPLALPSPALMPVAPLSPALVLPAPLSPALVLPVLPSPPPRPAA 414
Score = 32.3 bits (72), Expect = 0.20
Identities = 19/77 (24%), Positives = 28/77 (35%)
Frame = -1
Query: 141 SPPTPRSSTPIPHPPRRSLPSPPSPSPSPSPSPSPSPSPRSTPIPHPRKRSPASPSPSPS 200
+P P P P +P + P +P P + P P +P +P P
Sbjct: 385 APEAPGPG*GAPEAPGPG*GAPLASGPGWGAPEAPGPGWGAPEAPGPGWGAPEAPGPG*G 206
Query: 201 LSKSPSPSESPSLAPSP 217
++P P LAP P
Sbjct: 205 APEAPGPG*GAPLAPGP 155
>TC87086 similar to SP|Q41001|BCP_PEA Blue copper protein precursor. [Garden
pea] {Pisum sativum}, partial (87%)
Length = 1219
Score = 75.5 bits (184), Expect = 2e-14
Identities = 57/176 (32%), Positives = 81/176 (45%), Gaps = 9/176 (5%)
Frame = +1
Query: 1 MSSSSPILLMFIFSIWMLISYSESTDYLVGDSENSWKFPLPTRHALTRWASNYQFIVGDT 60
M+ S+P++L F I + + + +T + VGD ++ W WAS+ F VGD+
Sbjct: 433 MTFSNPLILGFFLVINVAVP-TLATVHTVGD-KSGWAIGSD----YNTWASDKTFAVGDS 594
Query: 61 ITFQYNNKTESVHEVEEEDYDRCGIRGEHVDHYDGNTMVVLKKTGIHHFISGKKRHCRLG 120
+ F Y +V EV+E DY C G T + LKK G H+FI HC G
Sbjct: 595 LVFNYG-AGHTVDEVKESDYKSCTTGNSISTDSSGPTTIPLKKAGTHYFICAVPGHCTGG 771
Query: 121 LKLAVVVMVAPVLSSPPP---------PPSPPTPRSSTPIPHPPRRSLPSPPSPSP 167
+KL+V V + SS P PS TP ++T P +S S S SP
Sbjct: 772 MKLSVKVKASSSASSAPSATPSPSGKGSPSDGTPAATTTTTTPSTQSASSSTSISP 939
Score = 30.4 bits (67), Expect = 0.75
Identities = 19/46 (41%), Positives = 27/46 (58%), Gaps = 1/46 (2%)
Frame = +1
Query: 198 SPSLSKSPSPSESPSLAPSPSDSV-ASLAPSSSPSDESPSPAPSPS 242
S S S +PS + SPS SPSD A+ +++PS +S S + S S
Sbjct: 799 SSSASSAPSATPSPSGKGSPSDGTPAATTTTTTPSTQSASSSTSIS 936
Score = 28.5 bits (62), Expect = 2.8
Identities = 16/50 (32%), Positives = 28/50 (56%)
Frame = +1
Query: 190 RSPASPSPSPSLSKSPSPSESPSLAPSPSDSVASLAPSSSPSDESPSPAP 239
++ +S S +PS + SPS SPS +P+ + + PS+ + S S +P
Sbjct: 793 KASSSASSAPSATPSPSGKGSPS-DGTPAATTTTTTPSTQSASSSTSISP 939
>TC76720 similar to GP|11121502|emb|CAC14888. putative extensin {Nicotiana
sylvestris}, partial (51%)
Length = 1290
Score = 74.7 bits (182), Expect = 3e-14
Identities = 48/132 (36%), Positives = 62/132 (46%)
Frame = +1
Query: 118 RLGLKLAVVVMVAPVLSSPPPPPSPPTPRSSTPIPHPPRRSLPSPPSPSPSPSPSPSPSP 177
R L ++ + + SS P P SS P P PSP + P+P+P+P+PSP
Sbjct: 499 RFSLVFLLLSFLVNIASSADSPAPTPATNSSLNSPSPTPIPTPSPANSPPAPTPTPTPSP 678
Query: 178 SPRSTPIPHPRKRSPASPSPSPSLSKSPSPSESPSLAPSPSDSVASLAPSSSPSDESPSP 237
S P P SP SPS S SPSPS SP APSP ++ + A S + E
Sbjct: 679 HSDSPPAP--------SPDNSPSSSPSPSPSSSP--APSPDEAADNNAISHTGIGEDGKS 828
Query: 238 APSPSSSGSKGG 249
+ SSG K G
Sbjct: 829 SGGGMSSGKKAG 864
Score = 69.3 bits (168), Expect = 1e-12
Identities = 44/108 (40%), Positives = 59/108 (53%)
Frame = +1
Query: 147 SSTPIPHPPRRSLPSPPSPSPSPSPSPSPSPSPRSTPIPHPRKRSPASPSPSPSLSKSPS 206
+ +P P P S + PSP+P P+PSP+ SP P TP P P SP S SP +PS
Sbjct: 553 ADSPAPTPATNSSLNSPSPTPIPTPSPANSP-PAPTPTPTP---SPHSDSPP-----APS 705
Query: 207 PSESPSLAPSPSDSVASLAPSSSPSDESPSPAPSPSSSGSKGGGAGHG 254
P SPS +PSPS S +P+ SP + + + A S + G G +G G
Sbjct: 706 PDNSPSSSPSPS---PSSSPAPSPDEAADNNAISHTGIGEDGKSSGGG 840
Score = 51.6 bits (122), Expect = 3e-07
Identities = 37/98 (37%), Positives = 42/98 (42%), Gaps = 15/98 (15%)
Frame = +2
Query: 135 SPPPP----PSPPTPRSSTPIPHPPRRSLPSPPSPSPSPSPSPSPSPSP---RSTPIPHP 187
SPPPP SPP P P PP SPP P P PSP P +S P P
Sbjct: 101 SPPPPVYKYKSPPPPVYKYKSPPPPVYKYKSPPPPPKKPYKYPSPPPPVYKYKSPPPPVY 280
Query: 188 RKRSPASP-----SPSPSLSK---SPSPSESPSLAPSP 217
+ +SP P SP P + K P P + P PSP
Sbjct: 281 KYKSPPPPVYKYKSPPPPVYKYKSPPPPPKKPYKYPSP 394
Score = 50.1 bits (118), Expect = 9e-07
Identities = 39/115 (33%), Positives = 43/115 (36%), Gaps = 8/115 (6%)
Frame = +2
Query: 135 SPPPPPSPPTPRSSTPIPHPPRRSLPSPPSP-----SPSPSPSPSPSPSP---RSTPIPH 186
SPPPPP P P P PP SPP P SP P SP P + P
Sbjct: 32 SPPPPPKKPY---KYPSPPPPVYKYKSPPPPVYKYKSPPPPVYKYKSPPPPVYKYKSPPP 202
Query: 187 PRKRSPASPSPSPSLSKSPSPSESPSLAPSPSDSVASLAPSSSPSDESPSPAPSP 241
P K+ PSP P + K SP SP V P + SP P P
Sbjct: 203 PPKKPYKYPSPPPPVYKYKSPPPPVYKYKSPPPPVYKYKSPPPPVYKYKSPPPPP 367
Score = 45.4 bits (106), Expect = 2e-05
Identities = 32/102 (31%), Positives = 41/102 (39%), Gaps = 2/102 (1%)
Frame = +2
Query: 140 PSPPTPRSSTPIPHPPRR--SLPSPPSPSPSPSPSPSPSPSPRSTPIPHPRKRSPASPSP 197
P PP + +P P PP++ PSPP P P P +S P P + +SP P P
Sbjct: 5 PPPPVYKYKSP-PPPPKKPYKYPSPPPPVYKYKSPPPPVYKYKSPPPPVYKYKSP--PPP 175
Query: 198 SPSLSKSPSPSESPSLAPSPSDSVASLAPSSSPSDESPSPAP 239
P P + P PSP V P + SP P
Sbjct: 176 VYKYKSPPPPPKKPYKYPSPPPPVYKYKSPPPPVYKYKSPPP 301
Score = 37.7 bits (86), Expect = 0.005
Identities = 26/70 (37%), Positives = 30/70 (42%), Gaps = 9/70 (12%)
Frame = +2
Query: 135 SPPPPP-------SPPTPRSSTPIPHPPRRSLPSPPSPSPSPSPSPSPSPSPRSTPIPHP 187
SPPPPP SPP P P PP SPP P P P +S P P P
Sbjct: 191 SPPPPPKKPYKYPSPPPPVYKYKSPPPPVYKYKSPPPPVYKYKSPPPPVYKYKSPP-PPP 367
Query: 188 RK--RSPASP 195
+K + P+ P
Sbjct: 368 KKPYKYPSPP 397
Score = 29.6 bits (65), Expect = 1.3
Identities = 22/70 (31%), Positives = 26/70 (36%)
Frame = +2
Query: 172 SPSPSPSPRSTPIPHPRKRSPASPSPSPSLSKSPSPSESPSLAPSPSDSVASLAPSSSPS 231
SP P +P P P K+ PSP P + K SP SP V P
Sbjct: 2 SPPPPVYKYKSP-PPPPKKPYKYPSPPPPVYKYKSPPPPVYKYKSPPPPVYKYKSPPPPV 178
Query: 232 DESPSPAPSP 241
+ SP P P
Sbjct: 179 YKYKSPPPPP 208
Score = 28.9 bits (63), Expect = 2.2
Identities = 17/45 (37%), Positives = 17/45 (37%)
Frame = +2
Query: 131 PVLSSPPPPPSPPTPRSSTPIPHPPRRSLPSPPSPSPSPSPSPSP 175
PV PPP P P PP SPP P P PSP
Sbjct: 272 PVYKYKSPPP----PVYKYKSPPPPVYKYKSPPPPPKKPYKYPSP 394
>TC80366 similar to GP|13877617|gb|AAK43886.1 protein kinase-like protein
{Arabidopsis thaliana}, partial (16%)
Length = 814
Score = 74.3 bits (181), Expect = 5e-14
Identities = 52/140 (37%), Positives = 69/140 (49%), Gaps = 9/140 (6%)
Frame = +3
Query: 135 SPPPPPSPPTPRSSTPIPHPPRRSLPSPPSPSPSPSPSPSPSPSPRSTPIPHPRKRSPAS 194
S PPP +P +P STP PP PS P PS +PS SP P +TP P P++
Sbjct: 3 SSPPPATPSSPPPSTPANPPP--VTPSAPPPSTPATPS---SPPPSTTPSAPP----PST 155
Query: 195 PSPSPSLSKSPSPSESPSLAPSPSDSVASLAPSSSP--SDESPSPA-------PSPSSSG 245
PS SP PSP +P+++P PS +P SD+SPSP PSP SS
Sbjct: 156 PSNSP-----PSPPTTPAISPPSGGGTTPSPPSRTPPSSDDSPSPPSSKTPPPPSPPSSS 320
Query: 246 SKGGGAGHGFLEVSIAMMMF 265
S G G ++ +++F
Sbjct: 321 SISTGTVIGIAVGAVVVLVF 380
Score = 66.6 bits (161), Expect = 9e-12
Identities = 38/97 (39%), Positives = 48/97 (49%), Gaps = 2/97 (2%)
Frame = +3
Query: 131 PVLSSPPPPPSPPTPRSSTP--IPHPPRRSLPSPPSPSPSPSPSPSPSPSPRSTPIPHPR 188
PV S PPP +P TP S P P P S PS PSP +P+ SP +TP P R
Sbjct: 63 PVTPSAPPPSTPATPSSPPPSTTPSAPPPSTPSNSPPSPPTTPAISPPSGGGTTPSPPSR 242
Query: 189 KRSPASPSPSPSLSKSPSPSESPSLAPSPSDSVASLA 225
+ SPSP SK+P P PS + + +V +A
Sbjct: 243 TPPSSDDSPSPPSSKTPPPPSPPSSSSISTGTVIGIA 353
Score = 33.1 bits (74), Expect = 0.12
Identities = 28/91 (30%), Positives = 34/91 (36%), Gaps = 6/91 (6%)
Frame = +3
Query: 136 PPPPPSPPT------PRSSTPIPHPPRRSLPSPPSPSPSPSPSPSPSPSPRSTPIPHPRK 189
PPP P P+ HPP + S P P P+P P PS + P P P
Sbjct: 465 PPPAQRPKVEPYGGPPQQWQNNAHPPSDHVVSKP---PPPAPIPPRPPSHVAPPPPPPAF 635
Query: 190 RSPASPSPSPSLSKSPSPSESPSLAPSPSDS 220
S + S S P SP +A S S
Sbjct: 636 ISSSGGSGSNYSGGELLPPPSPGIAFSSGKS 728
Score = 32.0 bits (71), Expect = 0.26
Identities = 20/50 (40%), Positives = 23/50 (46%), Gaps = 6/50 (12%)
Frame = +3
Query: 132 VLSSPPPP-PSPPTPRSSTPIPHPPRRSLPSPPSPSPSPS-----PSPSP 175
V+S PPPP P PP P S P PP + S + S P PSP
Sbjct: 552 VVSKPPPPAPIPPRPPSHVAPPPPPPAFISSSGGSGSNYSGGELLPPPSP 701
Score = 31.2 bits (69), Expect = 0.44
Identities = 36/152 (23%), Positives = 49/152 (31%), Gaps = 33/152 (21%)
Frame = +3
Query: 130 APVLSSPPPPPSPPTPRS---STPI------------------------------PHPPR 156
+P S PPPPSPP+ S T I + +
Sbjct: 270 SPPSSKTPPPPSPPSSSSISTGTVIGIAVGAVVVLVFFSICCICFRKKKRRRRDEEYYGQ 449
Query: 157 RSLPSPPSPSPSPSPSPSPSPSPRSTPIPHPRKRSPASPSPSPSLSKSPSPSESPSLAPS 216
++ PP P+ P P P + HP S P P+ PS P
Sbjct: 450 QNYQQPP-PAQRPKVEPYGGPPQQWQNNAHPPSDHVVSKPPPPAPIPPRPPSHVAPPPPP 626
Query: 217 PSDSVASLAPSSSPSDESPSPAPSPSSSGSKG 248
P+ +S S+ S P PSP + S G
Sbjct: 627 PAFISSSGGSGSNYSGGELLPPPSPGIAFSSG 722
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.311 0.129 0.393
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 11,878,764
Number of Sequences: 36976
Number of extensions: 409572
Number of successful extensions: 41012
Number of sequences better than 10.0: 2155
Number of HSP's better than 10.0 without gapping: 7008
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 18664
length of query: 268
length of database: 9,014,727
effective HSP length: 94
effective length of query: 174
effective length of database: 5,538,983
effective search space: 963783042
effective search space used: 963783042
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.8 bits)
S2: 57 (26.6 bits)
Medicago: description of AC136953.6


