

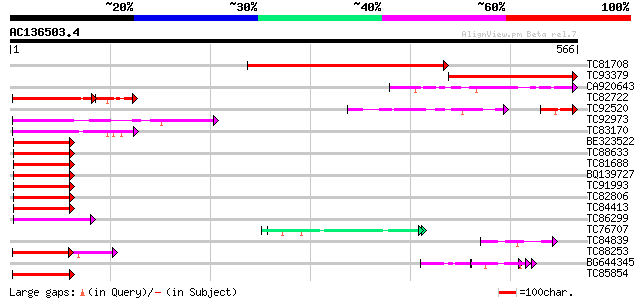
BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC136503.4 + phase: 0 /partial
(566 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC81708 similar to GP|12083536|gb|AAG48845.1 hypothetical protei... 429 e-120
TC93379 similar to GP|19569994|gb|AAL92295.1 hypothetical protei... 274 6e-74
CA920643 similar to GP|15010768|gb| AT3g06130/F28L1_7 {Arabidops... 122 3e-28
TC82722 similar to GP|15010768|gb|AAK74043.1 AT3g06130/F28L1_7 {... 100 1e-26
TC92520 similar to GP|15010768|gb|AAK74043.1 AT3g06130/F28L1_7 {... 68 6e-18
TC92973 weakly similar to GP|12328526|dbj|BAB21184. hypothetical... 81 9e-16
TC83170 similar to GP|15010768|gb|AAK74043.1 AT3g06130/F28L1_7 {... 80 3e-15
BE323522 similar to PIR|T05007|T05 hypothetical protein T19P19.9... 77 2e-14
TC88633 similar to PIR|T01827|T01827 hypothetical protein T15F16... 63 3e-10
TC81688 weakly similar to PIR|T51471|T51471 farnesylated protein... 62 5e-10
BQ139727 weakly similar to GP|21717170|gb hypothetical protein {... 62 5e-10
TC91993 weakly similar to GP|13899063|gb|AAK48953.1 Unknown prot... 62 6e-10
TC82806 similar to GP|4097573|gb|AAD09515.1| GMFP7 {Glycine max}... 62 8e-10
TC84413 similar to PIR|T51471|T51471 farnesylated protein ATFP6-... 60 2e-09
TC86299 similar to PIR|T50778|T50778 copper chaperone homolog CC... 60 2e-09
TC76707 similar to SP|Q07202|CORA_MEDSA Cold and drought-regulat... 60 3e-09
TC84839 similar to GP|19386827|dbj|BAB86205. hypothetical protei... 59 5e-09
TC88253 weakly similar to GP|15810597|gb|AAL07186.1 unknown prot... 58 9e-09
BG644345 weakly similar to GP|19697333|gb putative protein poten... 57 2e-08
TC85854 similar to PIR|T50778|T50778 copper chaperone homolog CC... 56 3e-08
>TC81708 similar to GP|12083536|gb|AAG48845.1 hypothetical protein {Oryza
sativa}, partial (6%)
Length = 605
Score = 429 bits (1102), Expect = e-120
Identities = 200/201 (99%), Positives = 200/201 (99%)
Frame = +1
Query: 238 GDIMDLPMQMHMKGKSGNYGEVKNGNGGGKKGGDDGGKKNKGEKQKGGGGDWGDEKNSSK 297
GDIMDLPMQMHMKGKSGNYGEVKNGNGGGKKGGDDGGKKNKGEKQKGGGGDWGDEKNSSK
Sbjct: 1 GDIMDLPMQMHMKGKSGNYGEVKNGNGGGKKGGDDGGKKNKGEKQKGGGGDWGDEKNSSK 180
Query: 298 KKNGKGKNGGGGFLVKFLGLGKKSKKGGGAADTTNKKKNNGGGGNSNNSKGKDGKKGEKL 357
KKNGKGKNGGGGFLVKFLGLGKKSKKGGGAADTTNKKKNNGGGGNSNNSKGKDGKKGEKL
Sbjct: 181 KKNGKGKNGGGGFLVKFLGLGKKSKKGGGAADTTNKKKNNGGGGNSNNSKGKDGKKGEKL 360
Query: 358 DKVEFDFQDFDITPHGKSGKGGNGNGNGKGAPAKGNANNGHGSNNNGNGGHMGPMGQMGA 417
DKVEFDFQDFDITPHGKSGKGGNGNGNGKGAPAKGNANNGHGSNNNGNGGHMGPMGQMGA
Sbjct: 361 DKVEFDFQDFDITPHGKSGKGGNGNGNGKGAPAKGNANNGHGSNNNGNGGHMGPMGQMGA 540
Query: 418 MNQMAAMNRMGPMGQMGSMDQ 438
MN MAAMNRMGPMGQMGSMDQ
Sbjct: 541 MNHMAAMNRMGPMGQMGSMDQ 603
Score = 39.3 bits (90), Expect = 0.004
Identities = 50/186 (26%), Positives = 67/186 (35%), Gaps = 5/186 (2%)
Frame = +1
Query: 43 GDVDPAKLLKKLKSSGKHAELWGGQKAMMINQNQNFQQQQP----QFKNMQIDNNKGGKN 98
G D K K K G + WG +K +N + +F + + KGG
Sbjct: 94 GGDDGGKKNKGEKQKGGGGD-WGDEKNSSKKKNGKGKNGGGGFLVKFLGLGKKSKKGGGA 270
Query: 99 NKPQNQKGQKGGVQVAQFQNPKGGKDMKVPNKSQKHVNFDLSEEEFDESDADFDDYDDED 158
N+K GG N GKD K E+ D+ + DF D+D
Sbjct: 271 ADTTNKKKNNGG---GGNSNNSKGKDGK-------------KGEKLDKVEFDFQDFDITP 402
Query: 159 DFDEDEEEEFGHGHGHGQGPQGNGFGQHPMHNNKMMMAMMQNQNGRGPQLGPGGGM-MMN 217
+ G+G+G G +GN H N NG G +GP G M MN
Sbjct: 403 HGKSGKGGN-GNGNGKGAPAKGNANNGHG-----------SNNNGNGGHMGPMGQMGAMN 546
Query: 218 GPAAMN 223
AAMN
Sbjct: 547 HMAAMN 564
>TC93379 similar to GP|19569994|gb|AAL92295.1 hypothetical protein
{Dictyostelium discoideum}, partial (1%)
Length = 535
Score = 274 bits (701), Expect = 6e-74
Identities = 128/128 (100%), Positives = 128/128 (100%)
Frame = +2
Query: 439 MRSMPAVQGLPAGAAAAAMNGGYYQGMQQMQPNPYHQQQQQHQQQQQQQQYMAMMMQQQQ 498
MRSMPAVQGLPAGAAAAAMNGGYYQGMQQMQPNPYHQQQQQHQQQQQQQQYMAMMMQQQQ
Sbjct: 2 MRSMPAVQGLPAGAAAAAMNGGYYQGMQQMQPNPYHQQQQQHQQQQQQQQYMAMMMQQQQ 181
Query: 499 QQQMQQQLQQQQMQQGNMNNMYPQHMMYGGGRPHPSMNYMGPPPMPSHPMADPITHVFSD 558
QQQMQQQLQQQQMQQGNMNNMYPQHMMYGGGRPHPSMNYMGPPPMPSHPMADPITHVFSD
Sbjct: 182 QQQMQQQLQQQQMQQGNMNNMYPQHMMYGGGRPHPSMNYMGPPPMPSHPMADPITHVFSD 361
Query: 559 ENTESCSI 566
ENTESCSI
Sbjct: 362 ENTESCSI 385
>CA920643 similar to GP|15010768|gb| AT3g06130/F28L1_7 {Arabidopsis
thaliana}, partial (15%)
Length = 776
Score = 122 bits (307), Expect = 3e-28
Identities = 83/198 (41%), Positives = 99/198 (49%), Gaps = 11/198 (5%)
Frame = -1
Query: 380 NGNGNGKGAPAKGNANNGHGSNNN------GNGGHMGPMGQMGAMNQMAAMNRMGPMGQM 433
NG G P G N NN GGH P G N + M+ PM QM
Sbjct: 767 NGGKKGMSIPVAGGVGNVQAMNNGFQKMMGAGGGHPHP-AMDGGGNNVGPMSM--PMNQM 597
Query: 434 GSMDQMRSMPAVQGLPAGAAAAAMNGGYYQG-----MQQMQPNPYHQQQQQHQQQQQQQQ 488
G ++PAVQGLPAG GGY+QG + M NPY QQQQQHQQQQQQQQ
Sbjct: 596 GG-----NIPAVQGLPAG-------GGYFQGGGGPGAEMMSGNPYQQQQQQHQQQQQQQQ 453
Query: 489 YMAMMMQQQQQQQMQQQLQQQQMQQGNMNNMYPQHMMYGGGRPHPSMNYMGPPPMPSHPM 548
MA M QQ+ M + Q+ + Q MMY RP P++NY+ PP +P
Sbjct: 452 LMAAAMMNQQRAAMVENDQR-----------FQQPMMY--ARPPPAVNYIYPP--YPYPP 318
Query: 549 ADPITHVFSDENTESCSI 566
DP +H FSDENT SC+I
Sbjct: 317 PDPYSHAFSDENTSSCNI 264
>TC82722 similar to GP|15010768|gb|AAK74043.1 AT3g06130/F28L1_7 {Arabidopsis
thaliana}, partial (17%)
Length = 683
Score = 100 bits (249), Expect(2) = 1e-26
Identities = 50/84 (59%), Positives = 61/84 (72%)
Frame = +1
Query: 3 LLKVNIHCEGCEQKVKKLLQKIEGVYSVNIDAEQGKVLVTGDVDPAKLLKKLKSSGKHAE 62
+LKVNIHC+GC+ KVKK+LQKI+GV++ IDAEQGKV V+G+VDP L+KKL SGKHA+
Sbjct: 316 VLKVNIHCDGCKHKVKKILQKIDGVFTTEIDAEQGKVTVSGNVDPNVLIKKLAKSGKHAQ 495
Query: 63 LWGGQKAMMINQNQNFQQQQPQFK 86
LW K N N N QFK
Sbjct: 496 LWSVPKPNN-NNNNNQNNLVNQFK 564
Score = 38.1 bits (87), Expect(2) = 1e-26
Identities = 23/44 (52%), Positives = 27/44 (61%), Gaps = 2/44 (4%)
Frame = +2
Query: 86 KNMQIDNNKGG--KNNKPQNQKGQKGGVQVAQFQNPKGGKDMKV 127
KNMQIDN KGG NNK QNQ QKG PKGG+ +++
Sbjct: 563 KNMQIDNGKGGGNNNNKGQNQ-NQKGS---GNNNQPKGGQQIQL 682
>TC92520 similar to GP|15010768|gb|AAK74043.1 AT3g06130/F28L1_7 {Arabidopsis
thaliana}, partial (13%)
Length = 756
Score = 68.2 bits (165), Expect(2) = 6e-18
Identities = 61/167 (36%), Positives = 69/167 (40%), Gaps = 6/167 (3%)
Frame = -2
Query: 338 GGGGNSNNSKGKDGKKGEKLDKVEFDFQDFDITPHGKSGKGGNGNGNGKGAPAKGNANNG 397
GGGGN GKK + G+SG G N NG G+ N G
Sbjct: 755 GGGGNGGIPNTNVGKK---------------VNGMGESGVQGMIN-NGLPNMGGGHPNVG 624
Query: 398 HGSNNNGNGGHMGPMGQMGAMNQMAAMNRMGPMGQMGSMDQMRSMPAVQGLPA------G 451
H +NN +G MG G MG N M M QMG +MPAVQGLPA G
Sbjct: 623 HMVSNNMSGMPMGGGGAMG--------NNMAMMSQMGG-----NMPAVQGLPAAAMNGGG 483
Query: 452 AAAAAMNGGYYQGMQQMQPNPYHQQQQQHQQQQQQQQYMAMMMQQQQ 498
+ GGY M NPY QQQQYMA MM QQ+
Sbjct: 482 GGGGGVCGGYGGPEMMMGGNPY-----------QQQQYMAAMMNQQR 375
Score = 40.8 bits (94), Expect(2) = 6e-18
Identities = 20/41 (48%), Positives = 26/41 (62%), Gaps = 5/41 (12%)
Frame = -3
Query: 531 PHPSMNYMGPPPM-----PSHPMADPITHVFSDENTESCSI 566
P ++NYM PPP P HP DP ++ F+DENT SCS+
Sbjct: 322 PPMAVNYMYPPPYSYPPPPQHPH-DPYSNFFNDENTSSCSV 203
>TC92973 weakly similar to GP|12328526|dbj|BAB21184. hypothetical
protein~similar to Arabidopsis thaliana chromosome 3
F28L1.7, partial (15%)
Length = 683
Score = 81.3 bits (199), Expect = 9e-16
Identities = 62/208 (29%), Positives = 89/208 (41%), Gaps = 2/208 (0%)
Frame = +2
Query: 3 LLKVNIHCEGCEQKVKKLLQKIEGVYSVNIDAEQGKVLVTGDVDPAKLLKKLKSSGKHAE 62
+LKV IHC+GC ++VKK+LQ IEGVY ID+ Q KV VTG+VD L+KKL SGK E
Sbjct: 137 VLKVLIHCDGCTKRVKKILQGIEGVYRTEIDSRQHKVTVTGNVDAETLIKKLSRSGKSVE 316
Query: 63 LWGGQKAMMINQNQNFQQQQPQFKNMQIDNNKGGKNNKPQNQKGQKGGVQVAQFQNPKGG 122
LW ++ P+ K+ + +KG N
Sbjct: 317 LW--------------PEKPPEKKDKKSSKSKGETENII--------------------N 394
Query: 123 KDMKVPNKSQKHVNFDLSEEEFDESDAD--FDDYDDEDDFDEDEEEEFGHGHGHGQGPQG 180
K++K K+ S E ES + DD D+ + D D +E + + G
Sbjct: 395 KEIKEDQKN--------STEHIGESSHEGCIDDAGDDGEEDSDHKEGNNGDKNNNKSEGG 550
Query: 181 NGFGQHPMHNNKMMMAMMQNQNGRGPQL 208
+ N A + NG G ++
Sbjct: 551 AKKKKKKKKKNXSGSASLTPNNGGGKEI 634
Score = 33.1 bits (74), Expect = 0.29
Identities = 32/114 (28%), Positives = 42/114 (36%), Gaps = 7/114 (6%)
Frame = +2
Query: 244 PMQMHMKGKSGNYGEVKNGNGGGKKGGDDGGKKNKGEKQKGG----GGDWGDEKNSSKKK 299
P + K S + GE +N K ++ GE G GD G+E + K+
Sbjct: 332 PPEKKDKKSSKSKGETENIINKEIKEDQKNSTEHIGESSHEGCIDDAGDDGEEDSDHKEG 511
Query: 300 NGKGKNGGGGFLVKFLGLGKKSKKGGGAADTTNKKKNNGGGGNS---NNSKGKD 350
N KN +K GGA KKK N G S NN GK+
Sbjct: 512 NNGDKNN--------------NKSEGGAKKKKKKKKKNXSGSASLTPNNGGGKE 631
Score = 30.4 bits (67), Expect = 1.9
Identities = 17/52 (32%), Positives = 27/52 (51%)
Frame = +2
Query: 326 GAADTTNKKKNNGGGGNSNNSKGKDGKKGEKLDKVEFDFQDFDITPHGKSGK 377
G D+ +K+ NNG + NN+K + G K +K K + +TP+ GK
Sbjct: 482 GEEDSDHKEGNNG---DKNNNKSEGGAKKKKKKKKKNXSGSASLTPNNGGGK 628
>TC83170 similar to GP|15010768|gb|AAK74043.1 AT3g06130/F28L1_7 {Arabidopsis
thaliana}, partial (21%)
Length = 935
Score = 79.7 bits (195), Expect = 3e-15
Identities = 65/153 (42%), Positives = 78/153 (50%), Gaps = 27/153 (17%)
Frame = +3
Query: 3 LLKVNIHCEGCE-QKVKK-LLQKIEGVYSVNIDAEQGKVLVTGDVDPAKLLKKLKSSGKH 60
+LKVNIH + +KVKK L + G + +G+ +G+VDP L+KKL SGKH
Sbjct: 354 VLKVNIHW*RLQSKKVKKNLAED*WGFCYLR*MLNKGR*PFSGNVDPNVLIKKLAKSGKH 533
Query: 61 AELWGGQKAMMINQNQNFQQQQP-QFKNMQIDNNKGG---KNNKPQ-----NQKGQKGG- 110
AELWG KA N N N Q P Q KNM IDN KGG NNK Q NQ KGG
Sbjct: 534 AELWGAPKA---NNNNNNQNNIPNQMKNMHIDNGKGGGGNNNNKGQKGPGNNQNQPKGGS 704
Query: 111 ---------------VQVAQFQNPKGGKDMKVP 128
Q+ Q Q+ KG D+KVP
Sbjct: 705 QQGQNPQQQQLQQQLQQLQQLQHIKGFGDLKVP 803
Score = 33.1 bits (74), Expect = 0.29
Identities = 27/78 (34%), Positives = 32/78 (40%)
Frame = +3
Query: 459 GGYYQGMQQMQPNPYHQQQQQHQQQQQQQQYMAMMMQQQQQQQMQQQLQQQQMQQGNMNN 518
GG + Q P + Q Q QQ Q QQQQ QQ QQLQQ Q +G +
Sbjct: 630 GGGGNNNNKGQKGPGNNQNQPKGGSQQGQN-----PQQQQLQQQLQQLQQLQHIKGFGDL 794
Query: 519 MYPQHMMYGGGRPHPSMN 536
PQ P P+ N
Sbjct: 795 KVPQFKDMNMKMPPPNQN 848
>BE323522 similar to PIR|T05007|T05 hypothetical protein T19P19.90 -
Arabidopsis thaliana, partial (87%)
Length = 599
Score = 77.0 bits (188), Expect = 2e-14
Identities = 37/61 (60%), Positives = 46/61 (74%)
Frame = +1
Query: 4 LKVNIHCEGCEQKVKKLLQKIEGVYSVNIDAEQGKVLVTGDVDPAKLLKKLKSSGKHAEL 63
LKV + C+GCE KVKK L + GV SV+I+ +Q KV VTG VDP K+LKK KS+GK AE+
Sbjct: 154 LKVRMDCDGCELKVKKTLSSLSGVQSVDINRKQQKVTVTGFVDPNKVLKKAKSTGKKAEI 333
Query: 64 W 64
W
Sbjct: 334 W 336
>TC88633 similar to PIR|T01827|T01827 hypothetical protein T15F16.6 -
Arabidopsis thaliana, partial (78%)
Length = 730
Score = 62.8 bits (151), Expect = 3e-10
Identities = 28/61 (45%), Positives = 42/61 (67%)
Frame = +2
Query: 4 LKVNIHCEGCEQKVKKLLQKIEGVYSVNIDAEQGKVLVTGDVDPAKLLKKLKSSGKHAEL 63
LK+ + CEGC +KVK +L ++G V++D +Q KV V+G V+P K+LK +S+ K EL
Sbjct: 158 LKIRMDCEGCARKVKHVLSGVKGAKKVDVDLKQQKVTVSGYVEPKKVLKAAQSTKKKVEL 337
Query: 64 W 64
W
Sbjct: 338 W 340
>TC81688 weakly similar to PIR|T51471|T51471 farnesylated protein ATFP6-like
protein - Arabidopsis thaliana, partial (44%)
Length = 697
Score = 62.4 bits (150), Expect = 5e-10
Identities = 25/61 (40%), Positives = 44/61 (71%)
Frame = +3
Query: 4 LKVNIHCEGCEQKVKKLLQKIEGVYSVNIDAEQGKVLVTGDVDPAKLLKKLKSSGKHAEL 63
++V + CEGCE+KVK ++ +GV S N+ +Q +V VTG +D ++L +++S+GK A++
Sbjct: 162 IRVKMDCEGCEKKVKNAVKDFDGVESYNVTKDQQRVTVTGHIDANEILDEVRSTGKTADM 341
Query: 64 W 64
W
Sbjct: 342 W 344
>BQ139727 weakly similar to GP|21717170|gb hypothetical protein {Oryza sativa
(japonica cultivar-group)}, partial (64%)
Length = 658
Score = 62.4 bits (150), Expect = 5e-10
Identities = 29/61 (47%), Positives = 41/61 (66%)
Frame = +2
Query: 4 LKVNIHCEGCEQKVKKLLQKIEGVYSVNIDAEQGKVLVTGDVDPAKLLKKLKSSGKHAEL 63
LKV + C GCE+ VK + K++G+ SV +D E +V VTG V+ K+LK ++ SGK AE
Sbjct: 254 LKVRMCCTGCERVVKNAIYKLKGIDSVKVDLEMERVTVTGYVERNKVLKAVRRSGKRAEF 433
Query: 64 W 64
W
Sbjct: 434 W 436
>TC91993 weakly similar to GP|13899063|gb|AAK48953.1 Unknown protein
{Arabidopsis thaliana}, partial (82%)
Length = 605
Score = 62.0 bits (149), Expect = 6e-10
Identities = 25/61 (40%), Positives = 45/61 (72%)
Frame = +1
Query: 4 LKVNIHCEGCEQKVKKLLQKIEGVYSVNIDAEQGKVLVTGDVDPAKLLKKLKSSGKHAEL 63
+KV + C+GCE++V+ + ++GV V ++ +Q KV V+G+VD ++LKK++++GK AE
Sbjct: 193 IKVKMDCDGCERRVRNSVAYMKGVKQVEVNRKQSKVTVSGNVDRNRVLKKIQNTGKRAEF 372
Query: 64 W 64
W
Sbjct: 373 W 375
>TC82806 similar to GP|4097573|gb|AAD09515.1| GMFP7 {Glycine max}, partial
(96%)
Length = 748
Score = 61.6 bits (148), Expect = 8e-10
Identities = 28/62 (45%), Positives = 44/62 (70%), Gaps = 1/62 (1%)
Frame = +3
Query: 4 LKVNIHCEGCEQKVKKLLQKIEGVYSVNIDAEQGKVLVTGDVDPAKLLKKLK-SSGKHAE 62
+KV + CEGCE+KVKK ++ ++GV V +D + KV VTG V+P+K++ ++ +GK E
Sbjct: 108 VKVKMDCEGCERKVKKSVEGMKGVTQVEVDRKASKVTVTGYVEPSKVVARMSHRTGKRVE 287
Query: 63 LW 64
LW
Sbjct: 288 LW 293
>TC84413 similar to PIR|T51471|T51471 farnesylated protein ATFP6-like
protein - Arabidopsis thaliana, partial (68%)
Length = 702
Score = 60.5 bits (145), Expect = 2e-09
Identities = 28/62 (45%), Positives = 44/62 (70%), Gaps = 1/62 (1%)
Frame = +3
Query: 4 LKVNIHCEGCEQKVKKLLQKIEGVYSVNIDAEQGKVLVTGDVDPAKLLKKLKSSG-KHAE 62
+KV + C+GCE++V+ + ++GV SV I+ +Q KV V G VDP +LK+++S+G K AE
Sbjct: 126 IKVKMDCDGCERRVRNAVATMKGVKSVEINRKQSKVTVNGFVDPNMVLKRVRSTGKKRAE 305
Query: 63 LW 64
W
Sbjct: 306 FW 311
>TC86299 similar to PIR|T50778|T50778 copper chaperone homolog CCH
[imported] - soybean, partial (56%)
Length = 667
Score = 60.1 bits (144), Expect = 2e-09
Identities = 30/82 (36%), Positives = 46/82 (55%)
Frame = +3
Query: 4 LKVNIHCEGCEQKVKKLLQKIEGVYSVNIDAEQGKVLVTGDVDPAKLLKKLKSSGKHAEL 63
LKV + CEGC VK++L K++GV S +ID ++ KV+V G+V+P +LK + +GK
Sbjct: 186 LKVGMSCEGCVGAVKRVLGKLDGVESYDIDLKEQKVVVKGNVEPDTVLKTVSKTGKPTAF 365
Query: 64 WGGQKAMMINQNQNFQQQQPQF 85
W + + PQF
Sbjct: 366 WEAEAPSETKAQ*LLPKHLPQF 431
>TC76707 similar to SP|Q07202|CORA_MEDSA Cold and drought-regulated protein
CORA. [Alfalfa] {Medicago sativa}, partial (87%)
Length = 780
Score = 59.7 bits (143), Expect = 3e-09
Identities = 53/171 (30%), Positives = 67/171 (38%), Gaps = 9/171 (5%)
Frame = +1
Query: 252 KSGNYGEVKNGNGGGKKGGD----DGGKKNKGEKQKGGGGDW----GDEKNSSKKKNGKG 303
K G YG N GGG GG GG N G GGG + G + NG G
Sbjct: 151 KYGGYGGGYNHGGGGYNGGGYNHGGGGYNNGGGGYNHGGGGYNNGGGGYNHGGGGYNGGG 330
Query: 304 KN-GGGGFLVKFLGLGKKSKKGGGAADTTNKKKNNGGGGNSNNSKGKDGKKGEKLDKVEF 362
N GGGG+ G G + GGG + GGG N G G + E
Sbjct: 331 YNHGGGGY---NHGGGGYNHGGGGYNGGGGHGGHGGGGYNGGGGHGGHGAAESVAVQTEE 501
Query: 363 DFQDFDITPHGKSGKGGNGNGNGKGAPAKGNANNGHGSNNNGNGGHMGPMG 413
+ + +G GG +G G G+ ++G G N+G GGH G G
Sbjct: 502 KTNEVNDAKYG----GGYNHGGGSYNHGGGSYHHGGGGYNHGGGGHGGHGG 642
Score = 42.0 bits (97), Expect = 6e-04
Identities = 42/159 (26%), Positives = 52/159 (32%)
Frame = +1
Query: 258 EVKNGNGGGKKGGDDGGKKNKGEKQKGGGGDWGDEKNSSKKKNGKGKNGGGGFLVKFLGL 317
E N K GG GG + G GGG + G G NGGGG+
Sbjct: 124 EKTNEVNDAKYGGYGGGYNHGGGGYNGGGYNHG---------GGGYNNGGGGY------- 255
Query: 318 GKKSKKGGGAADTTNKKKNNGGGGNSNNSKGKDGKKGEKLDKVEFDFQDFDITPHGKSGK 377
N+GGGG +N G + HG G
Sbjct: 256 ------------------NHGGGGYNNGGGGYN---------------------HGGGGY 318
Query: 378 GGNGNGNGKGAPAKGNANNGHGSNNNGNGGHMGPMGQMG 416
G G +G G N+G G N+G GG+ G G G
Sbjct: 319 NGGGYNHGGGG-----YNHGGGGYNHGGGGYNGGGGHGG 420
>TC84839 similar to GP|19386827|dbj|BAB86205. hypothetical protein~similar
to Arabidopsis thaliana chromosome 5 T6I14_10, partial
(20%)
Length = 819
Score = 58.9 bits (141), Expect = 5e-09
Identities = 38/82 (46%), Positives = 39/82 (47%), Gaps = 5/82 (6%)
Frame = +1
Query: 471 NPYHQQQQQHQQQQQQQQYMAMMMQQQQQQQMQQQ-----LQQQQMQQGNMNNMYPQHMM 525
NP HQQ QQHQQQQQQQQ QQ QQ Q QQQ QQQQ Q G
Sbjct: 169 NPQHQQHQQHQQQQQQQQ------QQYQQHQQQQQPPHFHHQQQQQQSG----------- 297
Query: 526 YGGGRPHPSMNYMGPPPMPSHP 547
P P + PPPMP P
Sbjct: 298 -----PGPDFHRGPPPPMPQQP 348
Score = 35.8 bits (81), Expect = 0.045
Identities = 29/100 (29%), Positives = 36/100 (36%), Gaps = 15/100 (15%)
Frame = +1
Query: 458 NGGYYQGMQQMQPNPYHQQQQQHQQQQQQQQYMAMMMQQQQQ---------------QQM 502
N + Q Q Q QQQ Q QQQQQ + QQQQ QQ
Sbjct: 169 NPQHQQHQQHQQQQQQQQQQYQQHQQQQQPPHFHHQQQQQQSGPGPDFHRGPPPPMPQQP 348
Query: 503 QQQLQQQQMQQGNMNNMYPQHMMYGGGRPHPSMNYMGPPP 542
++Q N+ + + GG P P GPPP
Sbjct: 349 PPMMRQPSASSTNLGSEF-----LPGGPPGPP----GPPP 441
>TC88253 weakly similar to GP|15810597|gb|AAL07186.1 unknown protein
{Arabidopsis thaliana}, partial (51%)
Length = 1122
Score = 58.2 bits (139), Expect = 9e-09
Identities = 32/63 (50%), Positives = 42/63 (65%), Gaps = 2/63 (3%)
Frame = +3
Query: 3 LLKVNIHCEGCEQKVKKLLQKIEGVYSVNIDAEQGKVLVTGD-VDPAKLLKKL-KSSGKH 60
+LKV++HCE C +KV K L+ EGV V D++ KV+V G DP K+LK+L K SGK
Sbjct: 213 VLKVDMHCEACARKVAKALKGFEGVEEVTADSKGSKVVVKGKAADPIKVLKRLQKKSGKK 392
Query: 61 AEL 63
EL
Sbjct: 393 VEL 401
Score = 55.1 bits (131), Expect = 7e-08
Identities = 30/108 (27%), Positives = 60/108 (54%), Gaps = 3/108 (2%)
Frame = +3
Query: 3 LLKVNIHCEGCEQKVKKLLQKIEGVYSVNIDAEQGKVLVTGDVDPAKLLKKL-KSSGKHA 61
+LK+ +HC+ C Q ++K ++KI+GV SV D + +V G +DP KL+ ++ K + K A
Sbjct: 507 VLKIRMHCDACAQVIQKRIRKIKGVESVETDLGNDQAIVKGVIDPTKLVDEVFKRTKKQA 686
Query: 62 ELWGGQ--KAMMINQNQNFQQQQPQFKNMQIDNNKGGKNNKPQNQKGQ 107
+ + K + + ++ + + K + NKG +NK + ++ +
Sbjct: 687 SIVKKEEKKEEEKKEEEKKEEVKEEEKKESEEENKGEDDNKTEIKRSE 830
>BG644345 weakly similar to GP|19697333|gb putative protein potential
transcriptional repressor Not4hp - Mus musculus, partial
(7%)
Length = 635
Score = 56.6 bits (135), Expect = 2e-08
Identities = 42/104 (40%), Positives = 49/104 (46%), Gaps = 1/104 (0%)
Frame = +1
Query: 411 PMGQMGAMNQMAAMNRMGPMGQMGSMDQMRSMPAVQGLPAGAAAAAMNGGYYQGMQQMQP 470
P Q ++ Q + P G Q +S P Q A+ G Q Q
Sbjct: 136 PQAQQPSLFQTPGQQQASPFQTPG---QQQSSP-FQTPGQQQASPFQTPGQQQPSPFSQI 303
Query: 471 NPYHQQQQQHQQQQQQQQYMAMMMQQQQQQ-QMQQQLQQQQMQQ 513
P+ QQQQQ Q QQQQQQ + QQQQQQ Q QQQLQ QQ QQ
Sbjct: 304 TPFSQQQQQLQFQQQQQQQQQQLQQQQQQQLQQQQQLQLQQQQQ 435
Score = 55.1 bits (131), Expect = 7e-08
Identities = 34/74 (45%), Positives = 39/74 (51%), Gaps = 9/74 (12%)
Frame = +1
Query: 462 YQGMQQMQPNPY--------HQQQQQHQQQQQQQQYMAMMMQQQQ-QQQMQQQLQQQQMQ 512
+Q Q QP+P+ QQQ Q QQQQQQQQ QQQQ QQQ Q QLQQQQ Q
Sbjct: 259 FQTPGQQQPSPFSQITPFSQQQQQLQFQQQQQQQQQQLQQQQQQQLQQQQQLQLQQQQQQ 438
Query: 513 QGNMNNMYPQHMMY 526
+ Q +Y
Sbjct: 439 LLQQHQSSQQQQLY 480
Score = 51.2 bits (121), Expect = 1e-06
Identities = 31/62 (50%), Positives = 36/62 (58%), Gaps = 2/62 (3%)
Frame = +1
Query: 461 YYQGMQQMQPNPYHQQQQQHQQQQQQQQYMAMMMQQQQQQQMQQQLQQ--QQMQQGNMNN 518
+ Q QQ+Q QQQQQ QQQQQQQ +QQQQQ Q+QQQ QQ QQ Q
Sbjct: 310 FSQQQQQLQFQQQQQQQQQQLQQQQQQQ-----LQQQQQLQLQQQQQQLLQQHQSSQQQQ 474
Query: 519 MY 520
+Y
Sbjct: 475 LY 480
>TC85854 similar to PIR|T50778|T50778 copper chaperone homolog CCH
[imported] - soybean, partial (81%)
Length = 853
Score = 56.2 bits (134), Expect = 3e-08
Identities = 25/62 (40%), Positives = 38/62 (60%)
Frame = +1
Query: 3 LLKVNIHCEGCEQKVKKLLQKIEGVYSVNIDAEQGKVLVTGDVDPAKLLKKLKSSGKHAE 62
+LKV + C GC V ++L+K+EGV S +ID ++ KV V G+V P + + +GK E
Sbjct: 100 VLKVKMSCSGCSGAVNRVLEKMEGVESFDIDMKEQKVTVKGNVKPQDVFDTVSKTGKKTE 279
Query: 63 LW 64
W
Sbjct: 280 FW 285
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.309 0.131 0.389
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 15,260,278
Number of Sequences: 36976
Number of extensions: 233619
Number of successful extensions: 8783
Number of sequences better than 10.0: 493
Number of HSP's better than 10.0 without gapping: 3246
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 5308
length of query: 566
length of database: 9,014,727
effective HSP length: 101
effective length of query: 465
effective length of database: 5,280,151
effective search space: 2455270215
effective search space used: 2455270215
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.1 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.7 bits)
S2: 61 (28.1 bits)
Medicago: description of AC136503.4


