

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
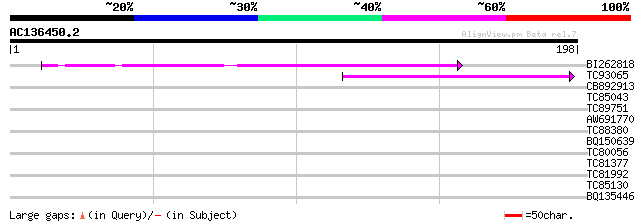
Query= AC136450.2 + phase: 0 /pseudo
(198 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
BI262818 weakly similar to GP|6642775|gb| gag-pol polyprotein {V... 46 8e-06
TC93065 44 3e-05
CB892913 similar to GP|1769897|emb lectin receptor kinase {Arabi... 32 0.12
TC85043 29 1.4
TC89751 similar to PIR|D84904|D84904 hypothetical protein At2g46... 28 1.8
AW691770 similar to PIR|B86473|B8 hypothetical protein AAG27107.... 28 2.3
TC88380 similar to GP|4324495|gb|AAD16897.1| glutamyl-tRNA reduc... 28 2.3
BQ150639 28 3.0
TC80056 similar to GP|12322859|gb|AAG51421.1 unknown protein co... 27 4.0
TC81377 phosphate transporter PHT2-1 [Medicago truncatula] 27 4.0
TC81992 weakly similar to PIR|T47841|T47841 hypothetical protein... 26 8.8
TC85130 similar to GP|6642643|gb|AAF20224.1| unknown protein {Ar... 26 8.8
BQ135446 similar to GP|537296|gb|AA chalcone reductase {Medicago... 26 8.8
>BI262818 weakly similar to GP|6642775|gb| gag-pol polyprotein {Vitis
vinifera}, partial (24%)
Length = 615
Score = 46.2 bits (108), Expect = 8e-06
Identities = 31/147 (21%), Positives = 72/147 (48%)
Frame = +1
Query: 12 VPKFNGDLEEFS*WKTNFYNHVMSLDEELWDILEGGIGDLVVDEEGADVDRRKHTPAQKK 71
+P F+G E + W ++ + D LW+ +E L + + + H + +
Sbjct: 175 LPVFDG--ENYHIWAARIEAYLEAND--LWEAVEEDYEVLPLSDNPTMAQIKNHKERKTR 342
Query: 72 LYKKHHTIRGALVKAIPKAEYMKMSDKSTAKSMFASLCANYEGSKKVREAKALMLVHQYE 131
K R L A+ + + ++ +A ++ L YEG +++R +AL L+ ++E
Sbjct: 343 KSKA----RATLFAAVSEEIFTRIMTIKSAFEIWNFLKTEYEGDERIRGMQALNLIREFE 510
Query: 132 LFKMKDGESIEQMYSRFHTLVSGLQIL 158
+ KMK+ E+I++ ++ ++ + +++L
Sbjct: 511 MQKMKESETIKEYANKLISIANKVRLL 591
>TC93065
Length = 783
Score = 44.3 bits (103), Expect = 3e-05
Identities = 22/81 (27%), Positives = 46/81 (56%)
Frame = +2
Query: 117 KVREAKALMLVHQYELFKMKDGESIEQMYSRFHTLVSGLQILKKSCVASDHVSKILRSLP 176
K +E ++ L ++E KMK+ E++ + R +V+ +++L + V KIL LP
Sbjct: 188 KNKENESAKLRREFEALKMKETETVREFSDRISKVVTQIRLLGEDLSDQRVVEKILVCLP 367
Query: 177 ARWRPKVTAIEETKDLNTLSV 197
+ K++++EE K+ + ++V
Sbjct: 368 EMFEAKISSLEENKNFSEITV 430
>CB892913 similar to GP|1769897|emb lectin receptor kinase {Arabidopsis
thaliana}, partial (35%)
Length = 837
Score = 32.3 bits (72), Expect = 0.12
Identities = 15/31 (48%), Positives = 22/31 (70%)
Frame = +1
Query: 168 VSKILRSLPARWRPKVTAIEETKDLNTLSVQ 198
+ KILRSL ++ V IEETKDL T++++
Sbjct: 559 MEKILRSLDPKFEHIVEKIEETKDLETMTIE 651
>TC85043
Length = 308
Score = 28.9 bits (63), Expect = 1.4
Identities = 17/44 (38%), Positives = 22/44 (49%)
Frame = +3
Query: 111 NYEGSKKVREAKALMLVHQYELFKMKDGESIEQMYSRFHTLVSG 154
NYEG KK + L L QY LF+ +I Q+Y F + G
Sbjct: 102 NYEGKKKNVKYSPLELERQYGLFESIVLPNIYQVYPMFFLIKKG 233
>TC89751 similar to PIR|D84904|D84904 hypothetical protein At2g46560
[imported] - Arabidopsis thaliana, partial (11%)
Length = 1446
Score = 28.5 bits (62), Expect = 1.8
Identities = 22/83 (26%), Positives = 38/83 (45%)
Frame = +1
Query: 65 HTPAQKKLYKKHHTIRGALVKAIPKAEYMKMSDKSTAKSMFASLCANYEGSKKVREAKAL 124
H+ Y K H + G L IPKA ++ +T + L + +G K+ +A++
Sbjct: 631 HSSLTSLNYDKDHNVDGMLWY-IPKAHSASVTKIATIPNTSLFLTGSTDGDVKLWDAESS 807
Query: 125 MLVHQYELFKMKDGESIEQMYSR 147
L+H + K+ D + Q SR
Sbjct: 808 KLLHHWS--KLHDKHTFLQSGSR 870
>AW691770 similar to PIR|B86473|B8 hypothetical protein AAG27107.1 [imported]
- Arabidopsis thaliana, partial (11%)
Length = 635
Score = 28.1 bits (61), Expect = 2.3
Identities = 18/46 (39%), Positives = 23/46 (49%)
Frame = +2
Query: 14 KFNGDLEEFS*WKTNFYNHVMSLDEELWDILEGGIGDLVVDEEGAD 59
KF G+L+EF+ NH SL L + GG+ V EEG D
Sbjct: 386 KFEGELQEFA-------NHAFSLRCVLECLQSGGVASDVKVEEGFD 502
>TC88380 similar to GP|4324495|gb|AAD16897.1| glutamyl-tRNA reductase
precursor {Glycine max}, partial (92%)
Length = 1689
Score = 28.1 bits (61), Expect = 2.3
Identities = 15/32 (46%), Positives = 20/32 (61%)
Frame = -3
Query: 146 SRFHTLVSGLQILKKSCVASDHVSKILRSLPA 177
S+ T VS L IL K+C+ S K+L SLP+
Sbjct: 1666 SKTETSVSRLNILFKACIFSSVSLKVLLSLPS 1571
>BQ150639
Length = 257
Score = 27.7 bits (60), Expect = 3.0
Identities = 12/34 (35%), Positives = 17/34 (49%)
Frame = +2
Query: 148 FHTLVSGLQILKKSCVASDHVSKILRSLPARWRP 181
FH L L +LK+ C ++D + S WRP
Sbjct: 29 FHVLEQELHLLKEPCDSNDQRGEKTPSYQGNWRP 130
>TC80056 similar to GP|12322859|gb|AAG51421.1 unknown protein contains
TNFR/NGFR cysteine-rich region; 28705-27186 {Arabidopsis
thaliana}, partial (98%)
Length = 879
Score = 27.3 bits (59), Expect = 4.0
Identities = 16/51 (31%), Positives = 28/51 (54%)
Frame = -2
Query: 146 SRFHTLVSGLQILKKSCVASDHVSKILRSLPARWRPKVTAIEETKDLNTLS 196
SRF SG L K+ + SKILR++ + + P+++ + K+ N+ S
Sbjct: 539 SRFWNFASGTFFL*KNDIIIGISSKILRNILSLYCPRISKAGQKKEQNSNS 387
>TC81377 phosphate transporter PHT2-1 [Medicago truncatula]
Length = 2236
Score = 27.3 bits (59), Expect = 4.0
Identities = 10/16 (62%), Positives = 14/16 (87%)
Frame = -1
Query: 147 RFHTLVSGLQILKKSC 162
RFH L+ GL++L+KSC
Sbjct: 1195 RFHKLMPGLELLEKSC 1148
>TC81992 weakly similar to PIR|T47841|T47841 hypothetical protein T2O9.150 -
Arabidopsis thaliana, partial (3%)
Length = 952
Score = 26.2 bits (56), Expect = 8.8
Identities = 19/80 (23%), Positives = 40/80 (49%)
Frame = +3
Query: 93 MKMSDKSTAKSMFASLCANYEGSKKVREAKALMLVHQYELFKMKDGESIEQMYSRFHTLV 152
+K K+ + SL Y+ + + + A+ L + + +MK+GESI ++R T+V
Sbjct: 660 LKQF*KTRHRRRSGSLKK*YQVTARGKRAQLQAL*RDW*ILQMKNGESIIDYFARTVTIV 839
Query: 153 SGLQILKKSCVASDHVSKIL 172
+ +++ + + KIL
Sbjct: 840 NKIRMHGEQMSNLTVIEKIL 899
>TC85130 similar to GP|6642643|gb|AAF20224.1| unknown protein {Arabidopsis
thaliana}, partial (37%)
Length = 775
Score = 26.2 bits (56), Expect = 8.8
Identities = 9/22 (40%), Positives = 15/22 (67%)
Frame = -2
Query: 155 LQILKKSCVASDHVSKILRSLP 176
L I + C+ S+H+ K+ +SLP
Sbjct: 582 LPIAQNGCIFSEHLQKVTKSLP 517
>BQ135446 similar to GP|537296|gb|AA chalcone reductase {Medicago sativa
subsp. sativa}, partial (14%)
Length = 718
Score = 26.2 bits (56), Expect = 8.8
Identities = 13/53 (24%), Positives = 31/53 (57%)
Frame = -1
Query: 130 YELFKMKDGESIEQMYSRFHTLVSGLQILKKSCVASDHVSKILRSLPARWRPK 182
+ +F++K G+ + YS++ ++L++ C+A+++ ++ S P RPK
Sbjct: 454 FPVFRVKVGDVLRIRYSQYSPY*---RVLRQFCIAANYKMSMIFSNPKFSRPK 305
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.319 0.134 0.389
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 5,448,237
Number of Sequences: 36976
Number of extensions: 63151
Number of successful extensions: 263
Number of sequences better than 10.0: 26
Number of HSP's better than 10.0 without gapping: 263
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 263
length of query: 198
length of database: 9,014,727
effective HSP length: 91
effective length of query: 107
effective length of database: 5,649,911
effective search space: 604540477
effective search space used: 604540477
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 56 (26.2 bits)
Medicago: description of AC136450.2


