

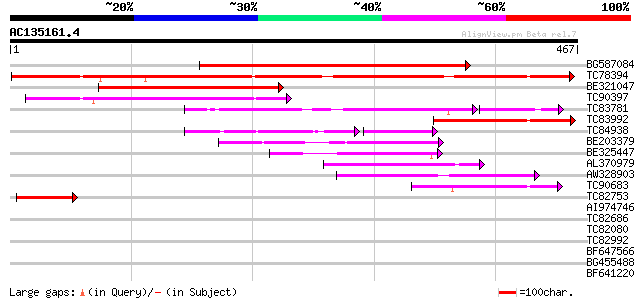
BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC135161.4 + phase: 0
(467 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
BG587084 homologue to GP|6002696|gb| histone acetyltransferase M... 429 e-120
TC78394 similar to GP|10177020|dbj|BAB10258. leucine zipper prot... 358 3e-99
BE321047 similar to GP|10177020|db leucine zipper protein {Arabi... 153 1e-37
TC90397 similar to GP|15010740|gb|AAK74029.1 AT5g59730/mth12_130... 129 2e-30
TC83781 homologue to PIR|S21495|S21495 tomato leucine zipper-con... 102 1e-29
TC83992 weakly similar to GP|10177020|dbj|BAB10258. leucine zipp... 105 3e-23
TC84938 weakly similar to GP|9758920|dbj|BAB09457.1 gb|AAD25781.... 81 3e-23
BE203379 similar to GP|22136732|gb unknown protein {Arabidopsis ... 85 7e-17
BE325447 weakly similar to GP|18700111|gb| At1g07000/F10K1_20 {A... 84 1e-16
AL370979 weakly similar to GP|15010740|gb| AT5g59730/mth12_130 {... 81 1e-15
AW328903 weakly similar to GP|8439895|gb|A It contains a interfe... 71 1e-12
TC90683 similar to GP|22136732|gb|AAM91685.1 unknown protein {Ar... 65 4e-11
TC82753 similar to GP|9758920|dbj|BAB09457.1 gb|AAD25781.1~gene_... 47 1e-05
AI974746 similar to GP|9955559|emb| putative protein {Arabidopsi... 40 0.001
TC82686 weakly similar to GP|10177020|dbj|BAB10258. leucine zipp... 39 0.004
TC82080 similar to GP|9757854|dbj|BAB08488.1 contains similarity... 34 0.10
TC82992 homologue to PIR|T05645|T05645 hypothetical protein F20D... 32 0.40
BF647566 similar to GP|9294624|dbj DNA repair protein RAD54-like... 32 0.68
BG455488 similar to GP|16604643|gb unknown protein {Arabidopsis ... 31 1.2
BF641220 weakly similar to PIR|G86203|G86 probable N-arginine di... 30 1.5
>BG587084 homologue to GP|6002696|gb| histone acetyltransferase MORF beta
{Homo sapiens}, partial (1%)
Length = 685
Score = 429 bits (1103), Expect = e-120
Identities = 214/223 (95%), Positives = 218/223 (96%)
Frame = +1
Query: 157 DLIPNFESLFCDQYSVSLRNELNTVLKKLGETIVGTLREFENTIRSKGPGNAPFFGGQLH 216
DLIPNFESLFCDQYSVSLRNELNTVLKKLGETIVGTLREFENTIRSKGPGNAPFFGGQLH
Sbjct: 1 DLIPNFESLFCDQYSVSLRNELNTVLKKLGETIVGTLREFENTIRSKGPGNAPFFGGQLH 180
Query: 217 PLVRFVMNFLTWICDYREILEQVFEDHGHVLLEYTKHDDTVPSSSSSSSSSSSSSSSLQM 276
PLVRFVMNFLTWICDYREILEQVFEDHGHVLLEYTKHDDTVPSSSSSSSSSSSSSSSLQM
Sbjct: 181 PLVRFVMNFLTWICDYREILEQVFEDHGHVLLEYTKHDDTVPSSSSSSSSSSSSSSSLQM 360
Query: 277 ERIMEVLESKLEAMFNIFNDPTLGYVYLMNSSRYIIIKTMENELGTLLGDGMLQRHSAKL 336
ERIMEVLESKLEAMFNIFNDPTLGYVYLMNSSRYIIIKTMENELGTLLGDGMLQRHSAKL
Sbjct: 361 ERIMEVLESKLEAMFNIFNDPTLGYVYLMNSSRYIIIKTMENELGTLLGDGMLQRHSAKL 540
Query: 337 RYNFEEYIRSSWGKVLEFLRLDNNLLVHPNMVGKSMKKQLKSF 379
RYNFEEYIRSSWGKVLEFLRLDNNLLVHPNMVG+ ++ + F
Sbjct: 541 RYNFEEYIRSSWGKVLEFLRLDNNLLVHPNMVGEVNEEAAQIF 669
Score = 28.1 bits (61), Expect = 7.5
Identities = 12/13 (92%), Positives = 13/13 (99%)
Frame = +2
Query: 368 VGKSMKKQLKSFN 380
+GKSMKKQLKSFN
Sbjct: 635 LGKSMKKQLKSFN 673
>TC78394 similar to GP|10177020|dbj|BAB10258. leucine zipper protein
{Arabidopsis thaliana}, partial (83%)
Length = 2109
Score = 358 bits (918), Expect = 3e-99
Identities = 209/470 (44%), Positives = 295/470 (62%), Gaps = 6/470 (1%)
Frame = +1
Query: 2 LSECSLEGTLMLEALSLETVNNLQETVKLMLNSGFNKECLIVYSSCRRECLEECLVKQFL 61
+++ ++ ++++AL TVN+L+E K M+ +GF KEC VY CRRE LEE L + L
Sbjct: 712 VAKAVVDYNVVIDALPPATVNDLREIAKRMIAAGFGKECSHVYGGCRREFLEESLSR--L 885
Query: 62 NSDNLTIKDVNM---EDLGLRIKRWIKAFKVAFKILFPTERQLCDIVFFEFS---AISDI 115
L+I +V+ +DL I+RWIKA VA KILFP+ER+LCD VF S A +D+
Sbjct: 886 GLQKLSISEVHKMQWQDLEDEIERWIKASNVALKILFPSERRLCDRVFSGLSSSSAAADL 1065
Query: 116 SFTDVCREFTIRLLNFPNVIANDQSNTTLLFRMLDMYETLHDLIPNFESLFCDQYSVSLR 175
SF +VCR I+LLNF + +A + LFR+LD++ET+ DLIP FESLF DQY L
Sbjct: 1066 SFMEVCRGSAIQLLNFSDAVAIGSRSPERLFRVLDVFETMRDLIPEFESLFSDQYCSFLV 1245
Query: 176 NELNTVLKKLGETIVGTLREFENTIRSKGPGNAPFFGGQLHPLVRFVMNFLTWICDYREI 235
N+ T K+LGE I GT E EN I S+ P A GG LHP+ R+VMN+L C +
Sbjct: 1246 NQAITNWKRLGEAITGTFMELENLI-SRDPVKAVVPGGGLHPITRYVMNYLRAACRSSKT 1422
Query: 236 LEQVFEDHGHVLLEYTKHDDTVPSSSSSSSSSSSSSSSLQMERIMEVLESKLEAMFNIFN 295
LE VF+D+ L +Y KHD+++ S+SS S+Q+ IM++LE LEA I+
Sbjct: 1423 LELVFKDNALSLKDYHKHDESL---------QSNSSFSVQISWIMDLLERNLEAKSRIYK 1575
Query: 296 DPTLGYVYLMNSSRYIIIKTMENELGTLLGDGMLQRHSAKLRYNFEEYIRSSWGKVLEFL 355
DP L V++MN+ RYI+ KT ++ELGTL+GD +++HS K+R Y RSSW K+L FL
Sbjct: 1576 DPALCSVFMMNNGRYIVQKTKDSELGTLMGDDWIRKHSTKVRQCHTNYQRSSWNKLLGFL 1755
Query: 356 RLDNNLLVHPNMVGKSMKKQLKSFNKLFNEICKAQSLWFIMDETLKEEIIVYLGENLLPA 415
+++ + K MK++LK FN F EIC+ QS WF+ DE LKEEI + + + LLPA
Sbjct: 1756 KVE-------TLAAKPMKEKLKMFNLHFEEICRVQSQWFVFDEQLKEEIRISIEKLLLPA 1914
Query: 416 YTNFIRKLHIVLKLEVKKPPDGYIEYETKDIKAILNNMFKIYRPSSCGRK 465
Y +FI + I+ E+ K D YI++ +DI+A LN++F+ S+ RK
Sbjct: 1915 YGSFIGRFQIL--PELAKNSDKYIKFGMEDIEARLNHLFQGSGGSNGSRK 2058
>BE321047 similar to GP|10177020|db leucine zipper protein {Arabidopsis
thaliana}, partial (25%)
Length = 495
Score = 153 bits (387), Expect = 1e-37
Identities = 77/152 (50%), Positives = 101/152 (65%)
Frame = +1
Query: 74 EDLGLRIKRWIKAFKVAFKILFPTERQLCDIVFFEFSAISDISFTDVCREFTIRLLNFPN 133
++L IKRWIK KVA +ILF +ER+LCD VFF S +D+SFTD+CRE ++LLNF
Sbjct: 40 KELEDEIKRWIKVSKVALRILFRSERRLCDQVFFGLSTTADLSFTDICRESMLQLLNFAE 219
Query: 134 VIANDQSNTTLLFRMLDMYETLHDLIPNFESLFCDQYSVSLRNELNTVLKKLGETIVGTL 193
IA + LFR+LDM+ET+ DLIP FESLF DQY+ S++NE T+ K+LGE I+G
Sbjct: 220 AIAIGSRSPERLFRVLDMFETMRDLIPEFESLFRDQYNGSMQNEATTIWKRLGEAIIGIF 399
Query: 194 REFENTIRSKGPGNAPFFGGQLHPLVRFVMNF 225
E EN I GG +HP+ +VMN+
Sbjct: 400 MELENLICHDPMNLEAVPGGGIHPITHYVMNY 495
>TC90397 similar to GP|15010740|gb|AAK74029.1 AT5g59730/mth12_130
{Arabidopsis thaliana}, partial (22%)
Length = 1162
Score = 129 bits (325), Expect = 2e-30
Identities = 76/223 (34%), Positives = 125/223 (55%), Gaps = 4/223 (1%)
Frame = +3
Query: 14 EALSLETVNNLQETVKLMLNSGFNKECLIVYSSCRRECLEECLVKQFLNSDNLT---IKD 70
E +S+ + +L+ M+N G+ KEC+ VY R+ ++E L L + LT I+
Sbjct: 426 ERVSMLAMADLKAIADCMINCGYGKECVKVYIVMRKSIVDEALYH--LGIERLTFSQIQK 599
Query: 71 VNMEDLGLRIKRWIKAFKVAFKILFPTERQLCDIVFFEFSA-ISDISFTDVCREFTIRLL 129
++ E + L+IK W+KA KVA + LF ER LCD VF I++ F ++ +E L
Sbjct: 600 MDWEVIELKIKTWLKAVKVAVRTLFHGERILCDDVFAAAGKRIAESCFAEITKEGATSLF 779
Query: 130 NFPNVIANDQSNTTLLFRMLDMYETLHDLIPNFESLFCDQYSVSLRNELNTVLKKLGETI 189
FP+++A + +FR LD+YE + D +S+F + + ++R + ++KL E +
Sbjct: 780 TFPDMVAKCKKTPEKMFRTLDLYEAISDHFQQIQSIFSFESTSNVRLQAINSMEKLAEAV 959
Query: 190 VGTLREFENTIRSKGPGNAPFFGGQLHPLVRFVMNFLTWICDY 232
L+EFE+ I+ K GG +HPL R+VMN+LT++ DY
Sbjct: 960 KTMLKEFESAIQ-KDSSKKQVSGGGVHPLTRYVMNYLTFLADY 1085
>TC83781 homologue to PIR|S21495|S21495 tomato leucine zipper-containing
protein - tomato, partial (5%)
Length = 1047
Score = 102 bits (255), Expect(2) = 1e-29
Identities = 79/244 (32%), Positives = 120/244 (48%), Gaps = 3/244 (1%)
Frame = +2
Query: 145 LFRMLDMYETLHDLIPNFESLFCDQYSVSLRNELNTVLKKLGETIVGTLREFEN-TIRSK 203
L +ML ++ETLH LI F L D S++ TV +LGE I + T R
Sbjct: 56 LSKMLAIFETLHHLISKFH-LGLDS---SVKEAAVTVQNRLGEAIRDLFLKLNYLTFRVP 223
Query: 204 GPGNAPFFGGQLHPLVRFVMNFLTWICDYREILEQVFEDHGHVLLEYTKHDDTVPSSSSS 263
G+ HP +++++T C R LEQV + EY K ++ V S
Sbjct: 224 AAKKVSRSDGRHHPTAVQIISYVTSACRSRHTLEQVLQ-------EYPKVNNGVVVKDSF 382
Query: 264 SSSSSSSSSSLQMERIMEVLESKLEAMFNIFNDPTLGYVYLMNSSRYIIIKTMENELGTL 323
QME IM++LE +L + + L Y+++MN+ R+I +L T+
Sbjct: 383 IE---------QMEWIMDMLEKRLTYKSKEYRELALRYLFMMNNRRHIEAMIKSLDLETI 535
Query: 324 LGDGMLQRHSAKLRYNFEEYIRSSWGKVLEFLRLDNN--LLVHPNMVGKSMKKQLKSFNK 381
G+ QR+ AK + + + Y R SW KVLEFL+LDNN ++ ++ +K++LK FNK
Sbjct: 536 FGNDWFQRNQAKFQQDLDLYQRYSWNKVLEFLKLDNNDCAALNGDVAEDILKEKLKLFNK 715
Query: 382 LFNE 385
F E
Sbjct: 716 QFEE 727
Score = 45.4 bits (106), Expect(2) = 1e-29
Identities = 26/69 (37%), Positives = 37/69 (52%)
Frame = +1
Query: 388 KAQSLWFIMDETLKEEIIVYLGENLLPAYTNFIRKLHIVLKLEVKKPPDGYIEYETKDIK 447
+ QS WF+ D+ LKEEII+ + LLP Y FI + L + + I Y +I+
Sbjct: 724 RVQSNWFVYDKKLKEEIIISVANTLLPVYGIFIGRFRDCLGIHANQ----CIRYGMFEIQ 891
Query: 448 AILNNMFKI 456
LNN+F I
Sbjct: 892 DRLNNLFLI 918
>TC83992 weakly similar to GP|10177020|dbj|BAB10258. leucine zipper protein
{Arabidopsis thaliana}, partial (14%)
Length = 615
Score = 105 bits (263), Expect = 3e-23
Identities = 56/117 (47%), Positives = 80/117 (67%)
Frame = +3
Query: 350 KVLEFLRLDNNLLVHPNMVGKSMKKQLKSFNKLFNEICKAQSLWFIMDETLKEEIIVYLG 409
KV FL+++NN + N V KSMK++LKSFN +F+++C+ QS WFI DE LKEEI + +
Sbjct: 6 KVFGFLKVENNGSMQQNGVAKSMKEKLKSFNMMFDDLCRVQSTWFIFDEQLKEEIRISIE 185
Query: 410 ENLLPAYTNFIRKLHIVLKLEVKKPPDGYIEYETKDIKAILNNMFKIYRPSSCGRKR 466
+ LLPAY NFI + V EV K D Y++Y T+DI+A LN++F+ S+ RK+
Sbjct: 186 KLLLPAYANFIARFQNV--AEVGKHADKYVKYGTEDIEAKLNDLFQGSSGSTGSRKQ 350
>TC84938 weakly similar to GP|9758920|dbj|BAB09457.1
gb|AAD25781.1~gene_id:MXI22.10~strong similarity to
unknown protein {Arabidopsis thaliana}, partial (28%)
Length = 728
Score = 80.9 bits (198), Expect(2) = 3e-23
Identities = 50/144 (34%), Positives = 74/144 (50%)
Frame = +1
Query: 145 LFRMLDMYETLHDLIPNFESLFCDQYSVSLRNELNTVLKKLGETIVGTLREFENTIRSKG 204
LFR+LDMYE L D +P E + D++ ++ E VL+ LGE + GT EFE+ IR++
Sbjct: 124 LFRILDMYEVLRDALPELEDMVTDEFVIT---EAKGVLRGLGEAVKGTFAEFESCIRNE- 291
Query: 205 PGNAPFFGGQLHPLVRFVMNFLTWICDYREILEQVFEDHGHVLLEYTKHDDTVPSSSSSS 264
P G +HPL R+VMN+L + DY ++ + E L + K+D S
Sbjct: 292 TSRRPVITGDVHPLPRYVMNYLKLLADYSNAMDSLLEISEEALYHF-KND----LGGDES 456
Query: 265 SSSSSSSSSLQMERIMEVLESKLE 288
+ S Q+ +M LE LE
Sbjct: 457 QLEALSPLGRQILLLMSELEHNLE 528
Score = 45.4 bits (106), Expect(2) = 3e-23
Identities = 19/61 (31%), Positives = 36/61 (58%)
Frame = +2
Query: 292 NIFNDPTLGYVYLMNSSRYIIIKTMENELGTLLGDGMLQRHSAKLRYNFEEYIRSSWGKV 351
N++ D L V+LMN+ Y++ K +++L +LGD +++ ++R Y+R+ W K
Sbjct: 539 NLYEDHALQQVFLMNNLHYLVRKVKDSDLIEVLGDNWVRKRRGQVRQYATGYLRACWSKA 718
Query: 352 L 352
L
Sbjct: 719 L 721
>BE203379 similar to GP|22136732|gb unknown protein {Arabidopsis thaliana},
partial (28%)
Length = 584
Score = 84.7 bits (208), Expect = 7e-17
Identities = 50/185 (27%), Positives = 92/185 (49%)
Frame = +1
Query: 173 SLRNELNTVLKKLGETIVGTLREFENTIRSKGPGNAPFFGGQLHPLVRFVMNFLTWICDY 232
++R+ + KKL +T T +FE + A G +HPL +V+N++ ++ DY
Sbjct: 19 AIRDAAMALTKKLAQTAQETFGDFEEAVEKDATKTA-VTDGTVHPLTSYVINYVKFLFDY 195
Query: 233 REILEQVFEDHGHVLLEYTKHDDTVPSSSSSSSSSSSSSSSLQMERIMEVLESKLEAMFN 292
R L+Q+F++ + SS +++ M RIM+ L+ L+
Sbjct: 196 RSTLKQLFQEF-------------------EGGNDSSQLATVTM-RIMQTLQINLDGKSK 315
Query: 293 IFNDPTLGYVYLMNSSRYIIIKTMENELGTLLGDGMLQRHSAKLRYNFEEYIRSSWGKVL 352
+ D L +++LMN+ YI+ +E LLGD +QRH ++ + +Y R++W K+L
Sbjct: 316 QYKDLALTHLFLMNNIHYIVRSVRRSEAKDLLGDDWVQRHRRIVQQHANQYKRNAWAKIL 495
Query: 353 EFLRL 357
+ L +
Sbjct: 496 QCLSI 510
>BE325447 weakly similar to GP|18700111|gb| At1g07000/F10K1_20 {Arabidopsis
thaliana}, partial (7%)
Length = 422
Score = 84.0 bits (206), Expect = 1e-16
Identities = 51/146 (34%), Positives = 79/146 (53%), Gaps = 4/146 (2%)
Frame = +3
Query: 215 LHPLVRFVMNFLTWICDYREILEQVFEDHGHVLLEYTKHDDTVPSSSSSSSSSSSSSSSL 274
L P+ + VMN+L +C ++ LEQVF D SS S
Sbjct: 6 LQPITQHVMNYLRVVCRSQQTLEQVFYD---------------------------SSLSS 104
Query: 275 QMERIMEVLESKLEAMFNIFNDPTLGYVYLMNSSRYIIIKTMENELGTLLGDGMLQRHSA 334
++ RI++ LES LEA + DP+LGY++L+N+ YI+ T +NELGTLLGD LQ+++
Sbjct: 105 KIHRIIDTLESNLEAKSKCYVDPSLGYIFLINNHTYIVEMTKDNELGTLLGDYWLQKYTE 284
Query: 335 KLRYNFEEYIRS----SWGKVLEFLR 356
K+ + +Y ++ + VL+F R
Sbjct: 285 KVWHYHRQYHKTKGAIASSVVLDFFR 362
>AL370979 weakly similar to GP|15010740|gb| AT5g59730/mth12_130 {Arabidopsis
thaliana}, partial (14%)
Length = 455
Score = 80.9 bits (198), Expect = 1e-15
Identities = 45/133 (33%), Positives = 73/133 (54%)
Frame = +2
Query: 259 SSSSSSSSSSSSSSSLQMERIMEVLESKLEAMFNIFNDPTLGYVYLMNSSRYIIIKTMEN 318
+SSSSSSSSSSS S ++ ++ VL KL+ + D L Y++L N+ +Y+++K +
Sbjct: 62 NSSSSSSSSSSSEISEKIAWLILVLLCKLDTKAEFYKDVALSYLFLANNMQYVVVKVRRS 241
Query: 319 ELGTLLGDGMLQRHSAKLRYNFEEYIRSSWGKVLEFLRLDNNLLVHPNMVGKSMKKQLKS 378
LG LLG+ L H K++ ++++ W KVL L + N + + +K
Sbjct: 242 NLGFLLGEEWLTNHELKVKEYVNKFVQIGWNKVLSTLPENENSTAEKTV--EQVKAIFVK 415
Query: 379 FNKLFNEICKAQS 391
FN F+E CK Q+
Sbjct: 416 FNAAFDEECKKQT 454
>AW328903 weakly similar to GP|8439895|gb|A It contains a interferon
alpha/beta domain PF|00143. EST gb|N96176 comes from
this gene., partial (15%)
Length = 485
Score = 70.9 bits (172), Expect = 1e-12
Identities = 41/167 (24%), Positives = 79/167 (46%)
Frame = +3
Query: 270 SSSSLQMERIMEVLESKLEAMFNIFNDPTLGYVYLMNSSRYIIIKTMENELGTLLGDGML 329
S + ++ ++ V+ KL+ + D L Y++L N+ +Y+++K ++ L +LG+ L
Sbjct: 3 SEIAKRLSWLILVVLCKLDGKAEFYKDVALSYLFLANNMQYVVVKVRKSNLRFILGEDWL 182
Query: 330 QRHSAKLRYNFEEYIRSSWGKVLEFLRLDNNLLVHPNMVGKSMKKQLKSFNKLFNEICKA 389
+H K++ +Y R +W KVL + N + + + FN F+E +
Sbjct: 183 LKHEMKVKEYVTKYERMAWSKVLS--------SIPENPTVEKASENFQGFNVEFDEAFRM 338
Query: 390 QSLWFIMDETLKEEIIVYLGENLLPAYTNFIRKLHIVLKLEVKKPPD 436
Q LW + D L+ EI L ++ Y F K + L ++ P+
Sbjct: 339 QYLWVVPDLELRNEIKESLVSKIVFKYREFYVKFRVGLDSVIRYSPE 479
>TC90683 similar to GP|22136732|gb|AAM91685.1 unknown protein {Arabidopsis
thaliana}, partial (19%)
Length = 554
Score = 65.5 bits (158), Expect = 4e-11
Identities = 37/129 (28%), Positives = 69/129 (52%), Gaps = 5/129 (3%)
Frame = +3
Query: 332 HSAKLRYNFEEYIRSSWGKVLEFLRLDNNLLV-----HPNMVGKSMKKQLKSFNKLFNEI 386
H ++ + +Y R SW K+L+ L + + N+ S+K++ K+FN F E+
Sbjct: 6 HRRTVQQHANQYKRISWAKILQRLTVQGGNSSGGGDSNRNISRSSVKERFKTFNTQFEEL 185
Query: 387 CKAQSLWFIMDETLKEEIIVYLGENLLPAYTNFIRKLHIVLKLEVKKPPDGYIEYETKDI 446
+ QS W + D L+E + + + E LLPAY +F+++ + +E K P YI Y +++
Sbjct: 186 HQRQSQWTVPDSELRESLRLAVAEVLLPAYRSFLKRFGPM--IENGKNPQKYIRYSPENL 359
Query: 447 KAILNNMFK 455
+ +L F+
Sbjct: 360 EQMLGEFFE 386
>TC82753 similar to GP|9758920|dbj|BAB09457.1
gb|AAD25781.1~gene_id:MXI22.10~strong similarity to
unknown protein {Arabidopsis thaliana}, partial (14%)
Length = 816
Score = 47.4 bits (111), Expect = 1e-05
Identities = 22/51 (43%), Positives = 34/51 (66%)
Frame = +1
Query: 6 SLEGTLMLEALSLETVNNLQETVKLMLNSGFNKECLIVYSSCRRECLEECL 56
SL L ++ + E++ NL++ + M+ SG+ +ECL VYSS RR+ L ECL
Sbjct: 604 SLGDDLFVDLVRPESILNLKDIIDRMVRSGYERECLQVYSSVRRDALVECL 756
>AI974746 similar to GP|9955559|emb| putative protein {Arabidopsis thaliana},
partial (13%)
Length = 498
Score = 40.4 bits (93), Expect = 0.001
Identities = 22/77 (28%), Positives = 42/77 (53%)
Frame = -1
Query: 390 QSLWFIMDETLKEEIIVYLGENLLPAYTNFIRKLHIVLKLEVKKPPDGYIEYETKDIKAI 449
+S W + DE L+ E+ V + ++PAY F+ + L+ + + YI+Y+ +DI+
Sbjct: 411 KSSWVVKDEQLQSELRVSVCGVVIPAYRAFVGR--FTQNLDSGRQVEKYIKYQPEDIETY 238
Query: 450 LNNMFKIYRPSSCGRKR 466
++ +F S GR+R
Sbjct: 237 IDELFDGKPHHSIGRRR 187
>TC82686 weakly similar to GP|10177020|dbj|BAB10258. leucine zipper protein
{Arabidopsis thaliana}, partial (5%)
Length = 858
Score = 38.9 bits (89), Expect = 0.004
Identities = 15/29 (51%), Positives = 25/29 (85%)
Frame = +1
Query: 12 MLEALSLETVNNLQETVKLMLNSGFNKEC 40
+++AL ET+N+L +TVK+M+++GF KEC
Sbjct: 763 IIDALPNETINHLHKTVKMMVDAGFEKEC 849
>TC82080 similar to GP|9757854|dbj|BAB08488.1 contains similarity to
apoptosis antagonizing transcription
factor~gene_id:MFB13.10, partial (37%)
Length = 908
Score = 34.3 bits (77), Expect = 0.10
Identities = 20/41 (48%), Positives = 27/41 (65%)
Frame = -3
Query: 256 TVPSSSSSSSSSSSSSSSLQMERIMEVLESKLEAMFNIFND 296
++PSSSSSSSSS+S SS ++ I SK ++F IF D
Sbjct: 162 SLPSSSSSSSSSTSLSSLTSLKDISSSYSSKSLSLFCIFLD 40
>TC82992 homologue to PIR|T05645|T05645 hypothetical protein F20D10.300 -
Arabidopsis thaliana, partial (7%)
Length = 354
Score = 32.3 bits (72), Expect = 0.40
Identities = 19/45 (42%), Positives = 27/45 (59%)
Frame = -2
Query: 257 VPSSSSSSSSSSSSSSSLQMERIMEVLESKLEAMFNIFNDPTLGY 301
+PSSSSSSSSSSSS SS I+ + S + N ++ T+ +
Sbjct: 155 LPSSSSSSSSSSSSYSSSSSSSIISIRASSSYSPPNPISNSTISF 21
>BF647566 similar to GP|9294624|dbj DNA repair protein RAD54-like
{Arabidopsis thaliana}, partial (12%)
Length = 669
Score = 31.6 bits (70), Expect = 0.68
Identities = 23/62 (37%), Positives = 33/62 (53%), Gaps = 3/62 (4%)
Frame = +3
Query: 241 EDHGHVLLEYTKHD-DTVPSSSSSSSSSSSSSSSLQMERIMEVLESKLEAMF--NIFNDP 297
ED VLL + D+ SSSSSSSSSS ++E++ + +EA+ N+F
Sbjct: 84 EDEDVVLLSSSSFSSDSDEEVEVSSSSSSSSSSDSEIEQVSQRKSQNVEALIKGNLFTTT 263
Query: 298 TL 299
TL
Sbjct: 264 TL 269
>BG455488 similar to GP|16604643|gb unknown protein {Arabidopsis thaliana},
partial (31%)
Length = 641
Score = 30.8 bits (68), Expect = 1.2
Identities = 26/69 (37%), Positives = 35/69 (50%), Gaps = 1/69 (1%)
Frame = +1
Query: 259 SSSSSSSSSSSSSSSLQMERIMEVLESKLEAMFNIFNDPTL-GYVYLMNSSRYIIIKTME 317
SS SSSSSSSSSSSS Q I E L ++ A + P L + SSR+I + ++
Sbjct: 55 SSPSSSSSSSSSSSSSQDILIRETL--RISAELASSSTPLLPSXTTFIGSSRFICCEQID 228
Query: 318 NELGTLLGD 326
+ D
Sbjct: 229 GRRWNYVAD 255
>BF641220 weakly similar to PIR|G86203|G86 probable N-arginine dibasic
convertase [imported] - Arabidopsis thaliana, partial
(5%)
Length = 634
Score = 30.4 bits (67), Expect = 1.5
Identities = 16/16 (100%), Positives = 16/16 (100%)
Frame = -2
Query: 258 PSSSSSSSSSSSSSSS 273
PSSSSSSSSSSSSSSS
Sbjct: 423 PSSSSSSSSSSSSSSS 376
Score = 29.3 bits (64), Expect = 3.4
Identities = 16/19 (84%), Positives = 17/19 (89%)
Frame = -2
Query: 259 SSSSSSSSSSSSSSSLQME 277
SSSSSSSSSSSSSSS +E
Sbjct: 408 SSSSSSSSSSSSSSSSSIE 352
Score = 27.7 bits (60), Expect = 9.8
Identities = 15/15 (100%), Positives = 15/15 (100%)
Frame = -2
Query: 259 SSSSSSSSSSSSSSS 273
SSSSSSSSSSSSSSS
Sbjct: 411 SSSSSSSSSSSSSSS 367
Score = 27.7 bits (60), Expect = 9.8
Identities = 15/15 (100%), Positives = 15/15 (100%)
Frame = -2
Query: 259 SSSSSSSSSSSSSSS 273
SSSSSSSSSSSSSSS
Sbjct: 414 SSSSSSSSSSSSSSS 370
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.322 0.138 0.400
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 15,284,622
Number of Sequences: 36976
Number of extensions: 251097
Number of successful extensions: 6455
Number of sequences better than 10.0: 128
Number of HSP's better than 10.0 without gapping: 2458
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 4382
length of query: 467
length of database: 9,014,727
effective HSP length: 100
effective length of query: 367
effective length of database: 5,317,127
effective search space: 1951385609
effective search space used: 1951385609
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 60 (27.7 bits)
Medicago: description of AC135161.4


