

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC135161.1 - phase: 0
(820 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
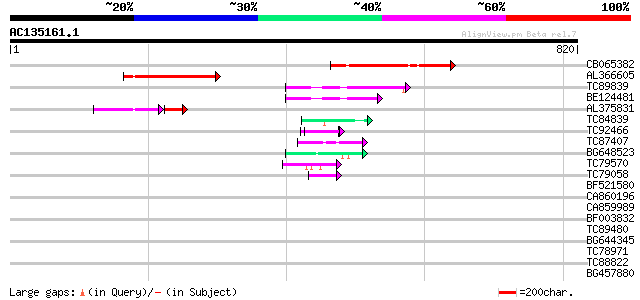
Score E
Sequences producing significant alignments: (bits) Value
CB065382 similar to PIR|T09671|T096 RPE15 protein - alfalfa (fra... 161 8e-40
AL366605 136 4e-32
TC89839 homologue to SP|P23466|CYAA_SACKL Adenylate cyclase (EC ... 117 2e-26
BE124481 96 7e-20
AL375831 74 3e-18
TC84839 similar to GP|19386827|dbj|BAB86205. hypothetical protei... 47 2e-05
TC92466 similar to GP|11141605|gb|AAG32022.1 LEUNIG {Arabidopsis... 47 3e-05
TC87407 similar to PIR|T49945|T49945 periaxin-like protein - Ara... 45 9e-05
BG648523 similar to GP|9955540|emb| putative protein {Arabidopsi... 44 3e-04
TC79570 similar to PIR|F86427|F86427 auxin response factor 6 (AR... 43 4e-04
TC79058 homologue to GP|20197423|gb|AAM15069.1 unknown protein {... 43 6e-04
BF521580 homologue to GP|20197423|gb|A unknown protein {Arabidop... 42 0.001
CA860196 weakly similar to GP|1654183|gb|A MYOD-like DNA-binding... 41 0.002
CA859989 similar to GP|18308125|gb bromodomain-containing protei... 41 0.002
BF003832 similar to GP|21524577|emb unnamed protein product {Ara... 41 0.002
TC89480 similar to GP|20260684|gb|AAM13240.1 unknown protein {Ar... 41 0.002
BG644345 weakly similar to GP|19697333|gb putative protein poten... 41 0.002
TC78971 similar to GP|20260346|gb|AAM13071.1 unknown protein {Ar... 40 0.003
TC88822 weakly similar to PIR|T06406|T06406 ripening protein E8 ... 39 0.006
BG457880 weakly similar to GP|1063689|gb|A ORF 66; this sequence... 39 0.008
>CB065382 similar to PIR|T09671|T096 RPE15 protein - alfalfa (fragment),
partial (9%)
Length = 624
Score = 161 bits (408), Expect = 8e-40
Identities = 86/182 (47%), Positives = 114/182 (62%)
Frame = -2
Query: 464 PQQNFQQQPYQQRPQQPRPPRMPINPIPVTYAELLPGLLKKNLVQTRTAPPIPEKLPSWY 523
P +N +QQR Q P P+ IP+ YAE LP LL + R P P+ LP +
Sbjct: 539 PPRNNHPPVHQQRQ*QQARPTFPL--IPMLYAE*LPTLLLRGHCTIRQGKPPPDPLPPRF 366
Query: 524 RLDQTCDFHEGGRGHNIETCYAFKSTVQRLINDGKITFTDSAPNVQTNPLPNHGAATVNM 583
R D CDFH+G GH++E CYA K V++LIN GK+TF ++ P+V NPLPNH A VNM
Sbjct: 365 RSDLKCDFHQGALGHDVEGCYALKHIVKKLINQGKLTFENNVPHVLDNPLPNH--AAVNM 192
Query: 584 IEDCQKTRPILNVQHIRTPLVPLHAKLCKVDLFEHDHDLCEICLMNSGGCQKVRNDIQGL 643
IE ++ P L+V+++ TPLVPLH KLC+ LF+HDH C+ N GC V+NDIQ L
Sbjct: 191 IEVYEEA-PGLDVRNVTTPLVPLHIKLCQASLFDHDHANCQE*FYNPLGCCVVQNDIQSL 15
Query: 644 LD 645
++
Sbjct: 14 MN 9
>AL366605
Length = 422
Score = 136 bits (342), Expect = 4e-32
Identities = 64/140 (45%), Positives = 93/140 (65%)
Frame = -3
Query: 165 DMKALREKGKFGKTAYDLCLVPSVQVPHKFKIPDFEKYKGSSCPEEHLKMYVRRMPAYAQ 224
++KA R KT DLCLV V VP KFKIPDF++Y G +CP+ H+ YVR+M Y
Sbjct: 417 EIKANRGNADSFKTQ-DLCLVSKVDVPKKFKIPDFDRYNGLTCPQNHIIKYVRKMGNYKD 241
Query: 225 DDQILIYYFQESLTGPASKWYTNLDKTRVQTFRDLCEAFVEQYSYNVDMTPDRSDLQAMT 284
+D ++I+ FQ+SL A++WYT+L K + TF +L AF Y +N + P+R L++++
Sbjct: 240 NDSLMIHCFQDSLMEDAAEWYTSLSKNDIHTFDELAAAFKSHYGFNTRLKPNREFLRSLS 61
Query: 285 QGDKETFKEYAQRWRDTAAQ 304
Q +E+F+EYAQRWR AA+
Sbjct: 60 QKKEESFREYAQRWRGAAAR 1
>TC89839 homologue to SP|P23466|CYAA_SACKL Adenylate cyclase (EC 4.6.1.1)
(ATP pyrophosphate-lyase) (Adenylyl cyclase). [Yeast],
partial (0%)
Length = 665
Score = 117 bits (292), Expect = 2e-26
Identities = 70/183 (38%), Positives = 87/183 (47%), Gaps = 3/183 (1%)
Frame = +3
Query: 400 IAAITPTSHPFQQTNNHPQIPQYPQIPQYPQFPQNPSPQNVQPQNVQQQNFQQQPYQQYP 459
+ +T TN+ P + Q PQ P P PQ P QQ P Q P
Sbjct: 180 VEKLTSLVSSLMATNDPPLVQQRPQSPYQPMCPQKPR--------------QQAPRQSIP 317
Query: 460 YQQYPQQNFQQQPYQQRPQQPRPPRMPINPIPVTYAELLPGLLKKNLVQTRTAPPIPEKL 519
Q PQ+ Q Q+ Q +PIPV YA+LLP LLKKNL+QT P +P L
Sbjct: 318 QNQVPQKFIPQNQVQKASQ--------CDPIPVKYADLLPILLKKNLIQTLPLPRVPNSL 473
Query: 520 PSWYRLDQTCDFHEGGRGHNIETCYAFKSTVQRLINDGKITFTDSAPNV---QTNPLPNH 576
P WYR D C FH+G GH+ E CY K VQ+LI + +F D V Q + P+
Sbjct: 474 PPWYRPDLNCVFHQGAPGHDTEQCYPLKEEVQKLIENNVWSFDDQDIKVLLQQQHLAPHS 653
Query: 577 GAA 579
GAA
Sbjct: 654 GAA 662
Score = 32.3 bits (72), Expect = 0.76
Identities = 21/71 (29%), Positives = 30/71 (41%), Gaps = 3/71 (4%)
Frame = +3
Query: 15 QTQAQAQIQAQAQALAQAQTQAQAVTE---AQAQSQAPPPPPPIRTQAEASSSWTLCADT 71
QTQ Q++ + L T +A E + S PP+ Q S +C
Sbjct: 105 QTQKMDQLEQEVHELRGEVTTLRAEVEKLTSLVSSLMATNDPPLVQQRPQSPYQPMCPQK 284
Query: 72 PRQSAPQRSAP 82
PRQ AP++S P
Sbjct: 285 PRQQAPRQSIP 317
>BE124481
Length = 538
Score = 95.5 bits (236), Expect = 7e-20
Identities = 54/140 (38%), Positives = 66/140 (46%)
Frame = +3
Query: 400 IAAITPTSHPFQQTNNHPQIPQYPQIPQYPQFPQNPSPQNVQPQNVQQQNFQQQPYQQYP 459
+ +T TN+ P + Q PQ P P PQ P QQ P Q P
Sbjct: 183 VEKLTSLVSSLMATNDPPLVQQRPQSPYQPMCPQKPR--------------QQAPRQSIP 320
Query: 460 YQQYPQQNFQQQPYQQRPQQPRPPRMPINPIPVTYAELLPGLLKKNLVQTRTAPPIPEKL 519
Q PQ+ Q Q+ Q +PIPV YA+LLP LLKKNL+QT P +P L
Sbjct: 321 QNQVPQKFIPQNQVQKASQ--------CDPIPVKYADLLPILLKKNLIQTLPLPRVPNSL 476
Query: 520 PSWYRLDQTCDFHEGGRGHN 539
P WYR D C FH+G GH+
Sbjct: 477 PPWYRPDLNCVFHQGAPGHD 536
Score = 30.4 bits (67), Expect = 2.9
Identities = 20/70 (28%), Positives = 29/70 (40%), Gaps = 3/70 (4%)
Frame = +3
Query: 16 TQAQAQIQAQAQALAQAQTQAQAVTE---AQAQSQAPPPPPPIRTQAEASSSWTLCADTP 72
TQ Q++ + L T +A E + S PP+ Q S +C P
Sbjct: 111 TQKMDQLEQEVHELRGEVTTLRAEVEKLTSLVSSLMATNDPPLVQQRPQSPYQPMCPQKP 290
Query: 73 RQSAPQRSAP 82
RQ AP++S P
Sbjct: 291 RQQAPRQSIP 320
>AL375831
Length = 467
Score = 73.9 bits (180), Expect(2) = 3e-18
Identities = 39/102 (38%), Positives = 59/102 (57%), Gaps = 1/102 (0%)
Frame = +2
Query: 122 TMTYSAPVIHTIPQTEEPIFHSGNVEAYEEVSDLREKY-DELRRDMKALREKGKFGKTAY 180
T + P++HT+PQ ++ + + ++ E+ ELR +++A R KT
Sbjct: 56 TAAVTEPLVHTLPQGININTQHRSIPVTKTMEEMMEELAKELRHEIQANRGNADSVKTQ- 232
Query: 181 DLCLVPSVQVPHKFKIPDFEKYKGSSCPEEHLKMYVRRMPAY 222
DLCLV V VP KFKIPDF++Y G +CP+ H+ YVR+M Y
Sbjct: 233 DLCLVSKVDVPKKFKIPDFDRYNGLTCPQNHIIKYVRKMGNY 358
Score = 36.6 bits (83), Expect(2) = 3e-18
Identities = 14/32 (43%), Positives = 23/32 (71%)
Frame = +3
Query: 225 DDQILIYYFQESLTGPASKWYTNLDKTRVQTF 256
+D ++I+ FQ+SL A++WYT+L K + TF
Sbjct: 366 NDSLMIHCFQDSLMEDAAEWYTSLSKNDIHTF 461
>TC84839 similar to GP|19386827|dbj|BAB86205. hypothetical protein~similar
to Arabidopsis thaliana chromosome 5 T6I14_10, partial
(20%)
Length = 819
Score = 47.4 bits (111), Expect = 2e-05
Identities = 37/107 (34%), Positives = 43/107 (39%), Gaps = 5/107 (4%)
Frame = +1
Query: 423 PQIPQYPQFPQNPSPQNVQPQNVQQQNFQQQP----YQQYPYQQYPQQNFQQQPYQQRPQ 478
P PQ+ Q Q+ Q Q Q QQ QQQP +QQ Q P +F + P PQ
Sbjct: 163 PHNPQHQQHQQHQQQQQQQQQQYQQHQQQQQPPHFHHQQQQQQSGPGPDFHRGPPPPMPQ 342
Query: 479 QPRP-PRMPINPIPVTYAELLPGLLKKNLVQTRTAPPIPEKLPSWYR 524
QP P R P +E LPG PP P P YR
Sbjct: 343 QPPPMMRQPSASSTNLGSEFLPG-----------GPPGPPGPPPHYR 450
Score = 33.1 bits (74), Expect = 0.44
Identities = 22/49 (44%), Positives = 26/49 (52%)
Frame = +1
Query: 435 PSPQNVQPQNVQQQNFQQQPYQQYPYQQYPQQNFQQQPYQQRPQQPRPP 483
P+P V N Q Q +QQ+ QQ QQ QQQ YQQ QQ +PP
Sbjct: 127 PTP*LVTMMYNDPHNPQHQQHQQHQQQQ--QQ--QQQQYQQHQQQQQPP 261
>TC92466 similar to GP|11141605|gb|AAG32022.1 LEUNIG {Arabidopsis thaliana},
partial (14%)
Length = 632
Score = 47.0 bits (110), Expect = 3e-05
Identities = 27/62 (43%), Positives = 32/62 (51%)
Frame = +1
Query: 421 QYPQIPQYPQFPQNPSPQNVQPQNVQQQNFQQQPYQQYPYQQYPQQNFQQQPYQQRPQQP 480
Q Q Q Q PQ Q+ Q Q Q QQ Q++ QQ QQ QQQP Q +PQQP
Sbjct: 373 QQQQQQQQQQQPQPQQSQHAQQQQQQHMQMQQLLMQRHAQQQQQQQQHQQQP-QSQPQQP 549
Query: 481 RP 482
+P
Sbjct: 550 QP 555
Score = 45.8 bits (107), Expect = 7e-05
Identities = 26/59 (44%), Positives = 33/59 (55%), Gaps = 1/59 (1%)
Frame = +1
Query: 427 QYPQFPQNPSPQNVQPQNVQQQNFQQQPYQQYPYQQYPQQNFQQQPYQQRPQ-QPRPPR 484
Q Q Q PQ Q Q+ QQQ Q QQ Q++ QQ QQQ +QQ+PQ QP+ P+
Sbjct: 376 QQQQQQQQQQPQPQQSQHAQQQQQQHMQMQQLLMQRHAQQQQQQQQHQQQPQSQPQQPQ 552
Score = 41.2 bits (95), Expect = 0.002
Identities = 23/59 (38%), Positives = 26/59 (43%)
Frame = +1
Query: 433 QNPSPQNVQPQNVQQQNFQQQPYQQYPYQQYPQQNFQQQPYQQRPQQPRPPRMPINPIP 491
Q Q QPQ Q Q+ QQQ Q QQ Q QQ QQ+ Q +P P P P
Sbjct: 379 QQQQQQQQQPQPQQSQHAQQQQQQHMQMQQLLMQRHAQQQQQQQQHQQQPQSQPQQPQP 555
Score = 36.2 bits (82), Expect = 0.052
Identities = 26/71 (36%), Positives = 32/71 (44%), Gaps = 5/71 (7%)
Frame = +1
Query: 411 QQTNNHPQIPQYPQIPQYPQFPQNPSPQNVQPQNV-----QQQNFQQQPYQQYPYQQYPQ 465
+Q Q Q PQ PQ Q Q Q++Q Q + QQ QQQ +QQ P Q Q
Sbjct: 370 EQQQQQQQQQQQPQ-PQQSQHAQQQQQQHMQMQQLLMQRHAQQQQQQQQHQQQPQSQPQQ 546
Query: 466 QNFQQQPYQQR 476
QQQ + R
Sbjct: 547 PQPQQQQNRDR 579
>TC87407 similar to PIR|T49945|T49945 periaxin-like protein - Arabidopsis
thaliana, partial (44%)
Length = 1559
Score = 45.4 bits (106), Expect = 9e-05
Identities = 24/101 (23%), Positives = 49/101 (47%)
Frame = +3
Query: 417 PQIPQYPQIPQYPQFPQNPSPQNVQPQNVQQQNFQQQPYQQYPYQQYPQQNFQQQPYQQR 476
P++P+ P++P+ P+FP+ P P+ + + + P + P + P+ N + P +
Sbjct: 372 PKLPELPKVPELPKFPELPKPELPKVPELSMPEIPKIP--ELPKPELPKLNAPELP---K 536
Query: 477 PQQPRPPRMPINPIPVTYAELLPGLLKKNLVQTRTAPPIPE 517
+QP+ P +P + +P P + K + P +PE
Sbjct: 537 LEQPKVPELPKHELPKVSELPKPDIPKVPELPKPELPKVPE 659
>BG648523 similar to GP|9955540|emb| putative protein {Arabidopsis thaliana},
partial (9%)
Length = 770
Score = 43.5 bits (101), Expect = 3e-04
Identities = 37/139 (26%), Positives = 52/139 (36%), Gaps = 21/139 (15%)
Frame = +3
Query: 400 IAAITPTSHPFQQTNNHPQIPQYPQIPQYPQFPQNPSPQNVQPQNVQQQNFQQQPYQQYP 459
+ P S P + Q +P P P P P + P QQQ FQQ P Q
Sbjct: 321 VGVAMPLSIPSFDGSQGEQKQPHPGSIGAPPLPPGPHPSLLNPN--QQQPFQQNPQQIPQ 494
Query: 460 YQQYPQQNFQQQPYQQRPQQ---------------PRPP-RMPIN-----PIPVTYAELL 498
+QQ QQ+ P Q PRPP +MP P+P ++ +
Sbjct: 495 HQQQLQQHMGPLPMPPNMPQIQHSSHSSMLPHQHLPRPPPQMPHGMPGSLPVPTSHPMPI 674
Query: 499 PGLLKKNLVQTRTAPPIPE 517
PG + + PP+P+
Sbjct: 675 PGPMGMQGTMNQMGPPMPQ 731
>TC79570 similar to PIR|F86427|F86427 auxin response factor 6 (ARF6)
[imported] - Arabidopsis thaliana, partial (15%)
Length = 1143
Score = 43.1 bits (100), Expect = 4e-04
Identities = 35/105 (33%), Positives = 45/105 (42%), Gaps = 20/105 (19%)
Frame = +2
Query: 395 PAYQHIAAITPTSHPFQQTNNHPQIPQYPQIPQ---YPQFPQN-----PSPQNVQPQNVQ 446
P+ QH A++ P +N + + Q + Q + FP+N PS Q Q Q
Sbjct: 347 PSKQHPASLLQFQQPQNFSNGNAALMQNQMLQQSQPHQAFPKNQESQHPSQSQAQTQQFQ 526
Query: 447 Q----------QNF--QQQPYQQYPYQQYPQQNFQQQPYQQRPQQ 479
Q QN+ QQQ QQ QQ QQ QQQ QQ QQ
Sbjct: 527 QLLQHQHSFTNQNYHMQQQQQQQQQQQQQQQQQHQQQQQQQSQQQ 661
Score = 42.0 bits (97), Expect = 0.001
Identities = 31/88 (35%), Positives = 39/88 (44%)
Frame = +2
Query: 384 MVTHGGPQQTYPAYQHIAAITPTSHPFQQTNNHPQIPQYPQIPQYPQFPQNPSPQNVQPQ 443
M+ P Q +P Q + HP Q + Q Q+ Q+ Q+ N QN Q
Sbjct: 434 MLQQSQPHQAFPKNQE------SQHPSQ---SQAQTQQFQQLLQHQHSFTN---QNYHMQ 577
Query: 444 NVQQQNFQQQPYQQYPYQQYPQQNFQQQ 471
QQQ QQQ QQ +QQ QQ QQQ
Sbjct: 578 QQQQQQQQQQQQQQQQHQQQQQQQSQQQ 661
Score = 40.8 bits (94), Expect = 0.002
Identities = 26/68 (38%), Positives = 33/68 (48%)
Frame = +2
Query: 409 PFQQTNNHPQIPQYPQIPQYPQFPQNPSPQNVQPQNVQQQNFQQQPYQQYPYQQYPQQNF 468
P Q + HP Q Q Q+ Q Q+ Q ++QQQ QQQ QQ QQ+ QQ
Sbjct: 467 PKNQESQHPSQSQ-AQTQQFQQLLQHQHSFTNQNYHMQQQQQQQQQQQQQQQQQHQQQQQ 643
Query: 469 QQQPYQQR 476
QQ QQ+
Sbjct: 644 QQSQQQQQ 667
Score = 31.2 bits (69), Expect = 1.7
Identities = 27/98 (27%), Positives = 33/98 (33%)
Frame = +2
Query: 374 FPRKKEQEVGMVTHGGPQQTYPAYQHIAAITPTSHPFQQTNNHPQIPQYPQIPQYPQFPQ 433
FP+ +E + + QQ QH H F N H
Sbjct: 464 FPKNQESQHPSQSQAQTQQFQQLLQH-------QHSFTNQNYH----------------- 571
Query: 434 NPSPQNVQPQNVQQQNFQQQPYQQYPYQQYPQQNFQQQ 471
+Q Q QQQ QQQ QQ+ QQ Q QQQ
Sbjct: 572 ------MQQQQQQQQQQQQQQQQQHQQQQQQQSQQQQQ 667
>TC79058 homologue to GP|20197423|gb|AAM15069.1 unknown protein {Arabidopsis
thaliana}, partial (71%)
Length = 627
Score = 42.7 bits (99), Expect = 6e-04
Identities = 23/51 (45%), Positives = 28/51 (54%), Gaps = 3/51 (5%)
Frame = +1
Query: 432 PQNPSPQNVQPQ-NVQQQNFQQQPY--QQYPYQQYPQQNFQQQPYQQRPQQ 479
PQ P++ P QQ + QQ Y Q YP Q YPQQ + Q Y Q+PQQ
Sbjct: 136 PQGYPPKDAYPPPGYPQQGYPQQGYPPQGYPQQGYPQQGYPPQQYAQQPQQ 288
Score = 37.7 bits (86), Expect = 0.018
Identities = 25/71 (35%), Positives = 28/71 (39%), Gaps = 2/71 (2%)
Frame = +1
Query: 408 HPFQQTNNHPQIPQYPQIPQYPQ--FPQNPSPQNVQPQNVQQQNFQQQPYQQYPYQQYPQ 465
H Q P YP YP +PQ PQ P Q P Q YP Q YP
Sbjct: 103 HQQQPPVGVPPPQGYPPKDAYPPPGYPQQGYPQQGYPP-------QGYPQQGYPQQGYPP 261
Query: 466 QNFQQQPYQQR 476
Q + QQP Q +
Sbjct: 262 QQYAQQPQQNK 294
Score = 35.0 bits (79), Expect = 0.12
Identities = 20/50 (40%), Positives = 24/50 (48%)
Frame = +1
Query: 435 PSPQNVQPQNVQQQNFQQQPYQQYPYQQYPQQNFQQQPYQQRPQQPRPPR 484
P PQ P++ P Q YP Q YP Q + QQ Y PQQ PP+
Sbjct: 130 PPPQGYPPKDAYPP--PGYPQQGYPQQGYPPQGYPQQGY---PQQGYPPQ 264
>BF521580 homologue to GP|20197423|gb|A unknown protein {Arabidopsis
thaliana}, partial (42%)
Length = 334
Score = 41.6 bits (96), Expect = 0.001
Identities = 21/47 (44%), Positives = 24/47 (50%), Gaps = 2/47 (4%)
Frame = +1
Query: 435 PSPQNVQPQNVQQQNFQQQPY--QQYPYQQYPQQNFQQQPYQQRPQQ 479
P P QQ + QQ Y Q YP Q YPQQ + Q Y Q+PQQ
Sbjct: 13 PPKDAYPPPGYPQQGYPQQGYPPQGYPQQGYPQQGYPPQQYAQQPQQ 153
Score = 37.7 bits (86), Expect = 0.018
Identities = 23/60 (38%), Positives = 26/60 (43%), Gaps = 2/60 (3%)
Frame = +1
Query: 419 IPQYPQIPQYPQ--FPQNPSPQNVQPQNVQQQNFQQQPYQQYPYQQYPQQNFQQQPYQQR 476
I YP YP +PQ PQ P Q P Q YP Q YP Q + QQP Q +
Sbjct: 1 IKGYPPKDAYPPPGYPQQGYPQQGYPP-------QGYPQQGYPQQGYPPQQYAQQPQQNK 159
Score = 33.1 bits (74), Expect = 0.44
Identities = 16/31 (51%), Positives = 18/31 (57%)
Frame = +1
Query: 454 PYQQYPYQQYPQQNFQQQPYQQRPQQPRPPR 484
P Q YP Q YP Q + QQ Y PQQ PP+
Sbjct: 46 PQQGYPQQGYPPQGYPQQGY---PQQGYPPQ 129
>CA860196 weakly similar to GP|1654183|gb|A MYOD-like DNA-binding protein
{Trichinella spiralis}, partial (7%)
Length = 398
Score = 41.2 bits (95), Expect = 0.002
Identities = 23/48 (47%), Positives = 26/48 (53%), Gaps = 3/48 (6%)
Frame = +1
Query: 442 PQNVQQQNFQQQPY---QQYPYQQYPQQNFQQQPYQQRPQQPRPPRMP 486
PQ QQ QQPY QQY Q PQQN + P Q PQ PP++P
Sbjct: 103 PQQYGQQQHVQQPYGQQQQYSQQYQPQQN--RPPLQMAPQSGPPPQVP 240
Score = 33.1 bits (74), Expect = 0.44
Identities = 24/49 (48%), Positives = 29/49 (58%), Gaps = 6/49 (12%)
Frame = +1
Query: 441 QPQNVQQQNFQQQPY---QQYPYQQYPQQNF-QQQPYQQR--PQQPRPP 483
+P + Q Q QQ Y QQY QQ+ QQ + QQQ Y Q+ PQQ RPP
Sbjct: 55 RPPHSQGQG-QQHNYGGPQQYGQQQHVQQPYGQQQQYSQQYQPQQNRPP 198
>CA859989 similar to GP|18308125|gb bromodomain-containing protein BRD4 long
variant {Mus musculus}, partial (1%)
Length = 375
Score = 41.2 bits (95), Expect = 0.002
Identities = 25/62 (40%), Positives = 31/62 (49%), Gaps = 15/62 (24%)
Frame = +3
Query: 437 PQNVQPQNVQQQNFQQQPYQQYPY--------QQYPQQNFQQQ-------PYQQRPQQPR 481
P ++ QNV QQ QQ P +Q Y QQ PQ + Q PYQQ+ QQP+
Sbjct: 24 PTSINTQNVTQQQQQQPPPEQQRYE*PASPYHQQRPQPTPEHQRYGQPAPPYQQQQQQPQ 203
Query: 482 PP 483
PP
Sbjct: 204 PP 209
>BF003832 similar to GP|21524577|emb unnamed protein product {Arabidopsis
thaliana}, partial (4%)
Length = 577
Score = 40.8 bits (94), Expect = 0.002
Identities = 43/147 (29%), Positives = 53/147 (35%), Gaps = 2/147 (1%)
Frame = +2
Query: 347 QLEEGVREGRLVKN--TTPASGTKKTGNHFPRKKEQEVGMVTHGGPQQTYPAYQHIAAIT 404
Q ++GV RLV N T SG HF + + EV T G +P ++
Sbjct: 20 QQQQGVVSSRLVSNFVTPTQSGQVAASPHFSHQYQAEVPPNTQKG----FPGSD--VSVL 181
Query: 405 PTSHPFQQTNNHPQIPQYPQIPQYPQFPQNPSPQNVQPQNVQQQNFQQQPYQQYPYQQYP 464
S FQQ NN+ Q P N +PQ+ Q N Q FQQ Q
Sbjct: 182 YNSQAFQQPNNNHHPFQQP----------NNNPQHFQQSNNNPQPFQQPNNSIALSSQVN 331
Query: 465 QQNFQQQPYQQRPQQPRPPRMPINPIP 491
N Q QP Q PI P
Sbjct: 332 SANPQHQPVMQYTADQVNSNPPIQQHP 412
Score = 28.9 bits (63), Expect = 8.3
Identities = 24/93 (25%), Positives = 35/93 (36%), Gaps = 2/93 (2%)
Frame = +2
Query: 403 ITPTSHPFQQTNNHPQIPQYPQIPQYPQ--FPQNPSPQNVQPQNVQQQNFQQQPYQQYPY 460
+TPT + H ++P Q FP + Q QQ N P+QQ
Sbjct: 65 VTPTQSGQVAASPHFSHQYQAEVPPNTQKGFPGSDVSVLYNSQAFQQPNNNHHPFQQ--- 235
Query: 461 QQYPQQNFQQQPYQQRPQQPRPPRMPINPIPVT 493
N Q +QQ P+P + P N I ++
Sbjct: 236 -----PNNNPQHFQQSNNNPQPFQQPNNSIALS 319
>TC89480 similar to GP|20260684|gb|AAM13240.1 unknown protein {Arabidopsis
thaliana}, partial (43%)
Length = 1068
Score = 40.8 bits (94), Expect = 0.002
Identities = 33/82 (40%), Positives = 40/82 (48%), Gaps = 6/82 (7%)
Frame = +1
Query: 404 TPTSHPFQQTNNHPQIPQYPQI--PQYPQF----PQNPSPQNVQPQNVQQQNFQQQPYQQ 457
TP S+P Q+ + Q Q P + Q QF Q P Q+ Q+ QQ QQQ YQQ
Sbjct: 94 TPNSNPQIQSQSQSQSNQDPNLYLQQQQQFLHQQQQQPFQQSQTTQSQFQQQQQQQLYQQ 273
Query: 458 YPYQQYPQQNFQQQPYQQRPQQ 479
Q+ QQ QQQ QQ QQ
Sbjct: 274 QQQQRILQQ--QQQQLQQPQQQ 333
>BG644345 weakly similar to GP|19697333|gb putative protein potential
transcriptional repressor Not4hp - Mus musculus, partial
(7%)
Length = 635
Score = 40.8 bits (94), Expect = 0.002
Identities = 28/68 (41%), Positives = 31/68 (45%)
Frame = +1
Query: 411 QQTNNHPQIPQYPQIPQYPQFPQNPSPQNVQPQNVQQQNFQQQPYQQYPYQQYPQQNFQQ 470
QQ + QI + Q Q QF Q Q Q Q QQQ QQQ QQ QQ QQ QQ
Sbjct: 277 QQPSPFSQITPFSQQQQQLQFQQQQQQQQQQLQQQQQQQLQQQ--QQLQLQQQQQQLLQQ 450
Query: 471 QPYQQRPQ 478
Q+ Q
Sbjct: 451 HQSSQQQQ 474
Score = 37.4 bits (85), Expect = 0.023
Identities = 31/90 (34%), Positives = 40/90 (44%), Gaps = 3/90 (3%)
Frame = +1
Query: 409 PFQQTNNHPQIPQYPQIPQYPQFPQNPSPQNVQPQNVQQQNFQQQPYQQYPYQQYPQQNF 468
PFQ T Q + QI + Q Q Q Q Q QQQ QQQ QQ QQ Q
Sbjct: 256 PFQ-TPGQQQPSPFSQITPFSQ--QQQQLQFQQQQQQQQQQLQQQQQQQLQQQQQLQLQQ 426
Query: 469 QQQPYQQRPQQPRPPRMPI---NPIPVTYA 495
QQQ Q+ Q + ++ + + P TY+
Sbjct: 427 QQQQLLQQHQSSQQQQLYLFTNDKTPATYS 516
Score = 35.8 bits (81), Expect = 0.068
Identities = 26/73 (35%), Positives = 30/73 (40%)
Frame = +1
Query: 407 SHPFQQTNNHPQIPQYPQIPQYPQFPQNPSPQNVQPQNVQQQNFQQQPYQQYPYQQYPQQ 466
S PFQ P Q P +P + Q Q +Q Q QQQ QQ QQ Q
Sbjct: 217 SSPFQTPGQQQASPFQTPGQQQPSPFSQITPFSQQQQQLQFQQQQQQQQQQLQQQQQQQL 396
Query: 467 NFQQQPYQQRPQQ 479
QQQ Q+ QQ
Sbjct: 397 QQQQQLQLQQQQQ 435
Score = 35.4 bits (80), Expect = 0.089
Identities = 32/85 (37%), Positives = 36/85 (41%)
Frame = +1
Query: 389 GPQQTYPAYQHIAAITPTSHPFQQTNNHPQIPQYPQIPQYPQFPQNPSPQNVQPQNVQQQ 448
G QQ P +Q P+ PF Q Q Q Q Q Q Q Q Q Q QQQ
Sbjct: 238 GQQQASP-FQTPGQQQPS--PFSQITPFSQQQQQLQFQQQQQQQQQQLQQQQQQQLQQQQ 408
Query: 449 NFQQQPYQQYPYQQYPQQNFQQQPY 473
Q Q QQ QQ+ Q + QQQ Y
Sbjct: 409 QLQLQQQQQQLLQQH-QSSQQQQLY 480
Score = 33.1 bits (74), Expect = 0.44
Identities = 27/74 (36%), Positives = 33/74 (44%), Gaps = 1/74 (1%)
Frame = +1
Query: 404 TPTSHPFQQTNNHPQIPQYPQIPQYPQF-PQNPSPQNVQPQNVQQQNFQQQPYQQYPYQQ 462
+P P QQ + Q P Q + Q P + Q +Q Q QQQ QQQ QQ Q
Sbjct: 220 SPFQTPGQQQASPFQTPGQQQPSPFSQITPFSQQQQQLQFQQQQQQQ-QQQLQQQQQQQL 396
Query: 463 YPQQNFQQQPYQQR 476
QQ Q Q QQ+
Sbjct: 397 QQQQQLQLQQQQQQ 438
>TC78971 similar to GP|20260346|gb|AAM13071.1 unknown protein {Arabidopsis
thaliana}, partial (48%)
Length = 2260
Score = 40.4 bits (93), Expect = 0.003
Identities = 26/53 (49%), Positives = 30/53 (56%), Gaps = 5/53 (9%)
Frame = +1
Query: 432 PQNPSPQNV--QPQNVQQQN-FQQQP--YQQYPYQQYPQQNFQQQPYQQRPQQ 479
PQ PQ QPQ QQQ+ FQQQ +Q Q PQQ FQQQ +Q + QQ
Sbjct: 250 PQQSQPQQTLFQPQQQQQQSPFQQQSSLFQPQQPQFQPQQQFQQQQFQAQQQQ 408
Score = 35.8 bits (81), Expect = 0.068
Identities = 23/55 (41%), Positives = 25/55 (44%)
Frame = +1
Query: 417 PQIPQYPQIPQYPQFPQNPSPQNVQPQNVQQQNFQQQPYQQYPYQQYPQQNFQQQ 471
PQ Q Q PQ Q SP Q Q Q Q QP QQ+ QQ+ Q QQQ
Sbjct: 250 PQQSQPQQTLFQPQQQQQQSPFQQQSSLFQPQQPQFQPQQQFQQQQFQAQQQQQQ 414
Score = 31.2 bits (69), Expect = 1.7
Identities = 24/59 (40%), Positives = 26/59 (43%)
Frame = +1
Query: 404 TPTSHPFQQTNNHPQIPQYPQIPQYPQFPQNPSPQNVQPQNVQQQNFQQQPYQQYPYQQ 462
TP QQT PQ Q Q P Q+ Q QPQ QQ FQQQ +Q QQ
Sbjct: 247 TPQQSQPQQTLFQPQQQQQ----QSPFQQQSSLFQPQQPQFQPQQQFQQQQFQAQQQQQ 411
>TC88822 weakly similar to PIR|T06406|T06406 ripening protein E8 homolog -
tomato, partial (31%)
Length = 1112
Score = 39.3 bits (90), Expect = 0.006
Identities = 23/72 (31%), Positives = 32/72 (43%)
Frame = -3
Query: 425 IPQYPQFPQNPSPQNVQPQNVQQQNFQQQPYQQYPYQQYPQQNFQQQPYQQRPQQPRPPR 484
+PQ P F +N +P Q Q+P QQ + P Q PY + P PR
Sbjct: 675 VPQSPSFHKNSTPSTFHVPIPNLQRATQKPLQQLRPHENP---IQPSPYD---KSPATPR 514
Query: 485 MPINPIPVTYAE 496
+ + P+P T AE
Sbjct: 513 LSVPPVPSTLAE 478
>BG457880 weakly similar to GP|1063689|gb|A ORF 66; this sequence overlaps at
the HindIII site with the sequence reported for the
OpMNPV lef-3, partial (5%)
Length = 669
Score = 38.9 bits (89), Expect = 0.008
Identities = 34/139 (24%), Positives = 48/139 (34%)
Frame = +3
Query: 377 KKEQEVGMVTHGGPQQTYPAYQHIAAITPTSHPFQQTNNHPQIPQYPQIPQYPQFPQNPS 436
+ Q+ + T P +Y H TP TN P YP + Q Q+P
Sbjct: 105 QNSQQPQVPTASDPNSSYYLTPHQPQQTPLGPSAPDTNQSP----YPNLQQPSQYPGGSV 272
Query: 437 PQNVQPQNVQQQNFQQQPYQQYPYQQYPQQNFQQQPYQQRPQQPRPPRMPINPIPVTYAE 496
QP Q QP Q Q P Q P +PQ P+P + P Y
Sbjct: 273 SAPSQPTPGQTPAQVAQPVQPV---QSPHMQHAQPPQPSQPQPPQPTQQAPPPQQQAY-- 437
Query: 497 LLPGLLKKNLVQTRTAPPI 515
+ ++ Q+ PP+
Sbjct: 438 -----WQPSMPQSHQQPPV 479
Score = 34.3 bits (77), Expect = 0.20
Identities = 35/129 (27%), Positives = 44/129 (33%), Gaps = 15/129 (11%)
Frame = +3
Query: 362 TPASGTKKTGNHFPRKKEQEVGMVTHGG---PQQTYPAY---QHIAAITPTSHPFQQTNN 415
TP + N P Q+ G P Q P Q + P P Q
Sbjct: 186 TPLGPSAPDTNQSPYPNLQQPSQYPGGSVSAPSQPTPGQTPAQVAQPVQPVQSPHMQHAQ 365
Query: 416 HPQIPQYPQIPQYPQFPQNPS---------PQNVQPQNVQQQNFQQQPYQQYPYQQYPQQ 466
PQ P PQ PQ Q P PQ+ Q V QN+ Y Q + PQQ
Sbjct: 366 PPQ-PSQPQPPQPTQQAPPPQQQAYWQPSMPQSHQQPPVAPQNWAYGGYTQDSFPSVPQQ 542
Query: 467 NFQQQPYQQ 475
+QP ++
Sbjct: 543 EPVKQPVKE 569
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.316 0.133 0.395
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 24,688,037
Number of Sequences: 36976
Number of extensions: 395557
Number of successful extensions: 5922
Number of sequences better than 10.0: 164
Number of HSP's better than 10.0 without gapping: 3657
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 5401
length of query: 820
length of database: 9,014,727
effective HSP length: 104
effective length of query: 716
effective length of database: 5,169,223
effective search space: 3701163668
effective search space used: 3701163668
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)
S2: 62 (28.5 bits)
Medicago: description of AC135161.1


