

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC133862.8 + phase: 0
(375 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
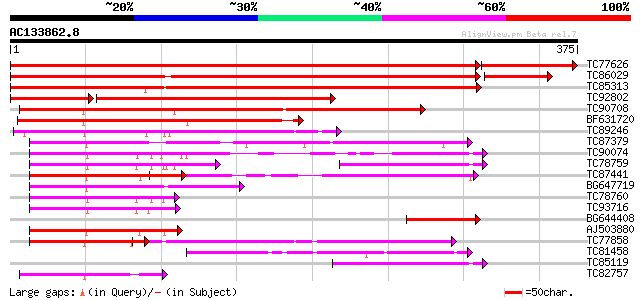
Score E
Sequences producing significant alignments: (bits) Value
TC77626 homologue to GP|5802244|gb|AAD51625.1| seed maturation p... 642 0.0
TC86029 homologue to GP|5802244|gb|AAD51625.1| seed maturation p... 592 e-170
TC85313 PIR|T09338|T09338 DnaJ-like protein MsJ1 - alfalfa, comp... 502 e-143
TC92802 homologue to PIR|T07371|T07371 dnaJ protein homolog - po... 292 e-105
TC90708 weakly similar to PIR|S26703|S26703 dnaJ protein homolog... 275 2e-74
BF631720 weakly similar to GP|10798648|em putative DNAJ protein ... 169 2e-42
TC89246 homologue to PIR|T06594|T06594 heat shock protein dnaJ -... 144 6e-35
TC87379 similar to PIR|T04618|T04618 heat shock protein homolog ... 139 2e-33
TC90074 similar to PIR|G84590|G84590 probable heat shock protein... 120 7e-28
TC78759 similar to GP|21618097|gb|AAM67147.1 putative heat shock... 106 2e-23
TC87441 similar to GP|21928031|gb|AAM78044.1 At3g62600/F26K9_30 ... 103 1e-22
BG647719 similar to GP|10177754|dbj DnaJ protein-like {Arabidops... 101 5e-22
TC78760 similar to PIR|T48161|T48161 heat shock protein 40-like ... 97 1e-20
TC93716 similar to GP|6179940|gb|AAF05720.1| DnaJ-like protein {... 94 1e-19
BG644408 homologue to PIR|T07371|T07 dnaJ protein homolog - pota... 93 1e-19
AJ503880 similar to GP|12838381|dbj DnaJ (Hsp40) homolog subfam... 93 2e-19
TC77858 similar to GP|20197886|gb|AAD22362.2 putative DnaJ prote... 92 2e-19
TC81458 similar to GP|21553367|gb|AAM62460.1 DnaJ protein-like {... 90 2e-18
TC85119 similar to PIR|G84590|G84590 probable heat shock protein... 77 8e-15
TC82757 similar to PIR|T08563|T08563 dnaJ-related protein T22F8.... 75 5e-14
>TC77626 homologue to GP|5802244|gb|AAD51625.1| seed maturation protein PM37
{Glycine max}, partial (98%)
Length = 1675
Score = 642 bits (1655), Expect = 0.0
Identities = 311/311 (100%), Positives = 311/311 (100%)
Frame = +1
Query: 1 MFGRAPKKSDNTKYYEILGVSKNASPDDLKKAYKKAAIKNHPDKGGDPEKFKELAQAYEV 60
MFGRAPKKSDNTKYYEILGVSKNASPDDLKKAYKKAAIKNHPDKGGDPEKFKELAQAYEV
Sbjct: 139 MFGRAPKKSDNTKYYEILGVSKNASPDDLKKAYKKAAIKNHPDKGGDPEKFKELAQAYEV 318
Query: 61 LSDPEKREVYDQYGEDALKEGMGGGGGGHDPFDIFSSFFGGGGFPGGGSSRGRRQRRGED 120
LSDPEKREVYDQYGEDALKEGMGGGGGGHDPFDIFSSFFGGGGFPGGGSSRGRRQRRGED
Sbjct: 319 LSDPEKREVYDQYGEDALKEGMGGGGGGHDPFDIFSSFFGGGGFPGGGSSRGRRQRRGED 498
Query: 121 VVHPLKVSLEDLYLGTSKKLSLSRNVLCSKCNGKGSKSGASMTCAGCQGSGMKISMRHLG 180
VVHPLKVSLEDLYLGTSKKLSLSRNVLCSKCNGKGSKSGASMTCAGCQGSGMKISMRHLG
Sbjct: 499 VVHPLKVSLEDLYLGTSKKLSLSRNVLCSKCNGKGSKSGASMTCAGCQGSGMKISMRHLG 678
Query: 181 ANMIQQMQHPCNECKGTGETISDKDRCPQCKGEKVVQQKKVLEVHVEKGMQNGQKITFPG 240
ANMIQQMQHPCNECKGTGETISDKDRCPQCKGEKVVQQKKVLEVHVEKGMQNGQKITFPG
Sbjct: 679 ANMIQQMQHPCNECKGTGETISDKDRCPQCKGEKVVQQKKVLEVHVEKGMQNGQKITFPG 858
Query: 241 EADEAPDTVTGDIVFVLQQKEHPKFKRKGEDLFVEHTLSLTEALCGFQFALTHLDSRQLL 300
EADEAPDTVTGDIVFVLQQKEHPKFKRKGEDLFVEHTLSLTEALCGFQFALTHLDSRQLL
Sbjct: 859 EADEAPDTVTGDIVFVLQQKEHPKFKRKGEDLFVEHTLSLTEALCGFQFALTHLDSRQLL 1038
Query: 301 IKSNPGEVVKP 311
IKSNPGEVVKP
Sbjct: 1039IKSNPGEVVKP 1071
Score = 130 bits (327), Expect = 8e-31
Identities = 63/63 (100%), Positives = 63/63 (100%)
Frame = +2
Query: 313 WMSVRRQHFMMSTLKKRREEGSKLSKRHMMRMMKCLVVLKEYNVLSSNEFGDILHGCVLA 372
WMSVRRQHFMMSTLKKRREEGSKLSKRHMMRMMKCLVVLKEYNVLSSNEFGDILHGCVLA
Sbjct: 1250 WMSVRRQHFMMSTLKKRREEGSKLSKRHMMRMMKCLVVLKEYNVLSSNEFGDILHGCVLA 1429
Query: 373 LVG 375
LVG
Sbjct: 1430 LVG 1438
>TC86029 homologue to GP|5802244|gb|AAD51625.1| seed maturation protein PM37
{Glycine max}, complete
Length = 1572
Score = 592 bits (1526), Expect = e-170
Identities = 289/312 (92%), Positives = 299/312 (95%), Gaps = 1/312 (0%)
Frame = +1
Query: 1 MFGRAPKKSDNTKYYEILGVSKNASPDDLKKAYKKAAIKNHPDKGGDPEKFKELAQAYEV 60
MFGRAPKKSD+T+YYEILGVSK AS DDLKKAYKKAAIKNHPDKGGDPEKFKELAQAYEV
Sbjct: 88 MFGRAPKKSDSTRYYEILGVSKTASQDDLKKAYKKAAIKNHPDKGGDPEKFKELAQAYEV 267
Query: 61 LSDPEKREVYDQYGEDALKEGMGGGGGG-HDPFDIFSSFFGGGGFPGGGSSRGRRQRRGE 119
LSDPEKRE+YD YGEDALKEGMGGGGGG HDPFDIFSSFFGGGG GGSSRGRRQRRGE
Sbjct: 268 LSDPEKREIYDTYGEDALKEGMGGGGGGGHDPFDIFSSFFGGGG---GGSSRGRRQRRGE 438
Query: 120 DVVHPLKVSLEDLYLGTSKKLSLSRNVLCSKCNGKGSKSGASMTCAGCQGSGMKISMRHL 179
DVVHPLKVSLEDLYLGTSKKLSLSRNVLCSKC+GKGSKSGASM CAGCQG+GMK+S+RHL
Sbjct: 439 DVVHPLKVSLEDLYLGTSKKLSLSRNVLCSKCSGKGSKSGASMKCAGCQGTGMKVSIRHL 618
Query: 180 GANMIQQMQHPCNECKGTGETISDKDRCPQCKGEKVVQQKKVLEVHVEKGMQNGQKITFP 239
G +MIQQMQHPCNECKGTGETI+DKDRCPQCKGEKVVQ+KKVLEVHVEKGMQN QKITFP
Sbjct: 619 GPSMIQQMQHPCNECKGTGETINDKDRCPQCKGEKVVQEKKVLEVHVEKGMQNSQKITFP 798
Query: 240 GEADEAPDTVTGDIVFVLQQKEHPKFKRKGEDLFVEHTLSLTEALCGFQFALTHLDSRQL 299
GEADEAPDTVTGDIVFVLQQKEHPKFKRK EDLFVEHTLSLTEALCGFQF LTHLD RQL
Sbjct: 799 GEADEAPDTVTGDIVFVLQQKEHPKFKRKSEDLFVEHTLSLTEALCGFQFVLTHLDGRQL 978
Query: 300 LIKSNPGEVVKP 311
LIKSNPGEVVKP
Sbjct: 979 LIKSNPGEVVKP 1014
Score = 45.4 bits (106), Expect = 3e-05
Identities = 25/46 (54%), Positives = 31/46 (67%), Gaps = 1/46 (2%)
Frame = +2
Query: 315 SVRRQHFMMSTLKKR-REEGSKLSKRHMMRMMKCLVVLKEYNVLSS 359
SVR+ HFMMS K+R S +++HMMRMM CLV +EYN SS
Sbjct: 1199 SVRKLHFMMSIWKRRIGGNNSNNNRKHMMRMMICLVGHREYNAPSS 1336
>TC85313 PIR|T09338|T09338 DnaJ-like protein MsJ1 - alfalfa, complete
Length = 1927
Score = 502 bits (1293), Expect = e-143
Identities = 240/315 (76%), Positives = 273/315 (86%), Gaps = 3/315 (0%)
Frame = +1
Query: 1 MFGRAP-KKSDNTKYYEILGVSKNASPDDLKKAYKKAAIKNHPDKGGDPEKFKELAQAYE 59
MFGR P +KSDNTKYY+ILGVSK+AS D++KKAY+KAA+KNHPDKGGDPEKFKEL QAYE
Sbjct: 280 MFGRGPTRKSDNTKYYDILGVSKSASEDEIKKAYRKAAMKNHPDKGGDPEKFKELGQAYE 459
Query: 60 VLSDPEKREVYDQYGEDALKEGMGGGGGG--HDPFDIFSSFFGGGGFPGGGSSRGRRQRR 117
VLSDPEK+E+YDQYGEDALKEGMGGG G H+PFDIF SFFG G F GGG SR RRQ++
Sbjct: 460 VLSDPEKKELYDQYGEDALKEGMGGGAGSSFHNPFDIFESFFGAG-FGGGGPSRARRQKQ 636
Query: 118 GEDVVHPLKVSLEDLYLGTSKKLSLSRNVLCSKCNGKGSKSGASMTCAGCQGSGMKISMR 177
GEDVVH +KVSLED+Y GT+KKLSLSRN LCSKC GKGSKSG + C GCQG+GMKI+ R
Sbjct: 637 GEDVVHSIKVSLEDVYNGTTKKLSLSRNALCSKCKGKGSKSGTAGRCFGCQGTGMKITRR 816
Query: 178 HLGANMIQQMQHPCNECKGTGETISDKDRCPQCKGEKVVQQKKVLEVHVEKGMQNGQKIT 237
+G MIQQMQH C +CKGTGE IS++DRCPQCKG K+ Q+KKVLEVHVEKGMQ G KI
Sbjct: 817 QIGLGMIQQMQHVCPDCKGTGEVISERDRCPQCKGNKITQEKKVLEVHVEKGMQQGHKIV 996
Query: 238 FPGEADEAPDTVTGDIVFVLQQKEHPKFKRKGEDLFVEHTLSLTEALCGFQFALTHLDSR 297
F G+ADEAPDT+TGDIVFVLQ K HPKF+R+ +DL +EH LSLTEALCGFQF +THLD R
Sbjct: 997 FEGQADEAPDTITGDIVFVLQVKGHPKFRRERDDLHIEHNLSLTEALCGFQFNVTHLDGR 1176
Query: 298 QLLIKSNPGEVVKPG 312
QLL+KSNPGEV+KPG
Sbjct: 1177QLLVKSNPGEVIKPG 1221
Score = 31.6 bits (70), Expect = 0.52
Identities = 21/53 (39%), Positives = 29/53 (54%), Gaps = 2/53 (3%)
Frame = +2
Query: 314 MSVRRQHFMMSTLKKRREEGSKLSKRHMMRMMKCLVVLKEY--NVLSSNEFGD 364
M VRR MMS L++R E S + RHMM MM ++ + N L+S+ D
Sbjct: 1406 MIVRRPPCMMSILQRR*VERSNNTARHMMTMMMKMMSTRSLGCNALNSSSIYD 1564
>TC92802 homologue to PIR|T07371|T07371 dnaJ protein homolog - potato,
partial (51%)
Length = 707
Score = 292 bits (748), Expect(2) = e-105
Identities = 140/159 (88%), Positives = 147/159 (92%), Gaps = 1/159 (0%)
Frame = +3
Query: 58 YEVLSDPEKREVYDQYGEDALKEGMGGGGGGHDPFDIFSSFFGGGGFPGGG-SSRGRRQR 116
YEVLSDPEKRE+YDQYGEDALKEGMGGGGGGHDPFDIFSSFFGG F GGG SSRGRRQR
Sbjct: 231 YEVLSDPEKREIYDQYGEDALKEGMGGGGGGHDPFDIFSSFFGGSPFGGGGGSSRGRRQR 410
Query: 117 RGEDVVHPLKVSLEDLYLGTSKKLSLSRNVLCSKCNGKGSKSGASMTCAGCQGSGMKISM 176
RGEDVVHPLKVSLEDLY GTSKKLSLSRNVLCSKC GKGSKSGASM C+GCQGSGMK+++
Sbjct: 411 RGEDVVHPLKVSLEDLYNGTSKKLSLSRNVLCSKCKGKGSKSGASMKCSGCQGSGMKVTI 590
Query: 177 RHLGANMIQQMQHPCNECKGTGETISDKDRCPQCKGEKV 215
R LG +MIQQMQHPCNECKGTGE I+DKD C QCKGEKV
Sbjct: 591 RQLGPSMIQQMQHPCNECKGTGEMINDKD*CGQCKGEKV 707
Score = 108 bits (270), Expect(2) = e-105
Identities = 50/55 (90%), Positives = 53/55 (95%)
Frame = +1
Query: 1 MFGRAPKKSDNTKYYEILGVSKNASPDDLKKAYKKAAIKNHPDKGGDPEKFKELA 55
MFGRAPKKSDNTKYYEILGV K A+P+DLKKAY+KAAIKNHPDKGGDPEKFKELA
Sbjct: 58 MFGRAPKKSDNTKYYEILGVPKTAAPEDLKKAYRKAAIKNHPDKGGDPEKFKELA 222
>TC90708 weakly similar to PIR|S26703|S26703 dnaJ protein homolog YDJ1 -
yeast (Saccharomyces cerevisiae), partial (43%)
Length = 1098
Score = 275 bits (703), Expect = 2e-74
Identities = 137/280 (48%), Positives = 175/280 (61%), Gaps = 11/280 (3%)
Frame = +2
Query: 7 KKSDNTKYYEILGVSKNASPDDLKKAYKKAAIKNHPDKGGD----PEKFKELAQAYEVLS 62
K TK Y+ LGV A+ +L+KAYK A+K HPDK EKFKE++ AYE+LS
Sbjct: 254 KMVKETKLYDTLGVKPEANDQELRKAYKTNALKYHPDKNAHNPEAEEKFKEISHAYEILS 433
Query: 63 DPEKREVYDQYGEDALKEGMGGGGGGHDPFDIFSSFFGGGGFPGG-------GSSRGRRQ 115
D +KR VYDQYGE L+ G GGGGGG D+F+ FFGGG F GG G R
Sbjct: 434 DSQKRAVYDQYGEAGLEGGAGGGGGGMPAEDLFAQFFGGGSFGGGLGGMFGGGGMPNRGP 613
Query: 116 RRGEDVVHPLKVSLEDLYLGTSKKLSLSRNVLCSKCNGKGSKSGASMTCAGCQGSGMKIS 175
+ + H KVSLED+Y G KL+L R+++CSKC G+G K GA C GC G GMK
Sbjct: 614 PKARTIHHTHKVSLEDVYRGKISKLALQRSIICSKCEGRGGKEGAVKRCTGCDGHGMKTM 793
Query: 176 MRHLGANMIQQMQHPCNECKGTGETISDKDRCPQCKGEKVVQQKKVLEVHVEKGMQNGQK 235
MR +G MIQ+ Q C +C G GE I DKDRC C G+K V +KVL VHV++G+++G K
Sbjct: 794 MRQMGP-MIQRFQTVCPDCNGEGEMIKDKDRCKHCSGKKTVVDRKVLHVHVDRGVRSGTK 970
Query: 236 ITFPGEADEAPDTVTGDIVFVLQQKEHPKFKRKGEDLFVE 275
+ F GE D+AP GD+VF ++QK H +F RK +DL +
Sbjct: 971 VEFRGEGDQAPGIQAGDVVFEIEQKPHTRFTRKDDDLLYQ 1090
>BF631720 weakly similar to GP|10798648|em putative DNAJ protein {Nicotiana
tabacum}, partial (25%)
Length = 630
Score = 169 bits (428), Expect = 2e-42
Identities = 93/197 (47%), Positives = 120/197 (60%), Gaps = 8/197 (4%)
Frame = +3
Query: 6 PKKSDNTKYYEILGVSKNASPDDLKKAYKKAAIKNHPDKGGD----PEKFKELAQAYEVL 61
P T+YY LGV NAS D++KKAY+K A+K HPDK E FK++++AYEVL
Sbjct: 42 PNMVRETEYYTRLGVQPNASQDEIKKAYRKLAVKYHPDKNPGNKEAEEMFKKISEAYEVL 221
Query: 62 SDPEKREVYDQYGEDALKEGMGGGGGGHDPFD-IFSSFFGGGGFPGGGSSRGRRQR--RG 118
SD +KR++YDQYGED LK G F+ +F F G GGF G G R R+ RG
Sbjct: 222 SDEQKRQMYDQYGEDGLKNSGYNPGDASSLFEHLFGGFGGFGGFGGFGGGRKRKSGPVRG 401
Query: 119 EDVVHPLKVSLEDLYLGTSKKLSLSRNVLCSKCNGKGS-KSGASMTCAGCQGSGMKISMR 177
+DVVHPL VSLEDLY G SKKL ++R++LC C G G+ K GA+ C C G G+++ +
Sbjct: 402 KDVVHPLTVSLEDLYKGVSKKLKITRHILCKTCTGSGAKKQGAATKCPTCDGRGIQVQDQ 581
Query: 178 HLGANMIQQMQHPCNEC 194
G QH N+C
Sbjct: 582 AHG-------QHESNKC 611
>TC89246 homologue to PIR|T06594|T06594 heat shock protein dnaJ - garden
pea, partial (50%)
Length = 1119
Score = 144 bits (363), Expect = 6e-35
Identities = 85/235 (36%), Positives = 125/235 (53%), Gaps = 18/235 (7%)
Frame = +1
Query: 3 GRAPKK-----SDNTKYYEILGVSKNASPDDLKKAYKKAAIKNHPDKGGDP---EKFKEL 54
G PK+ + ++ YY LGV K+A+ ++K AY++ A + HPD +P +KFKE+
Sbjct: 424 GTRPKRFHTVFAASSDYYSTLGVPKSATGKEIKAAYRRLARQYHPDVNKEPGATDKFKEI 603
Query: 55 AQAYEVLSDPEKREVYDQYGEDALKEGMGGGGGGH--DPFDIFSSFFGG--GGF-----P 105
+ AYEVLSD +KR +YDQYGE +K +GGG + +PFD+F +FFG GGF
Sbjct: 604 SNAYEVLSDDKKRALYDQYGEAGVKSSVGGGSSAYATNPFDLFETFFGPNMGGFSTMDPS 783
Query: 106 GGGSSRGRRQRRGEDVVHPLKVSLEDLYLGTSKKLSLSRNVLCSKCNGKGSKSGASM-TC 164
G G+ R +GED+ + + + GT K+ L C C G G+K G+ M C
Sbjct: 784 GFGTRRRSTVTKGEDIRYDFSLEFSEAIFGTEKEFELFHLETCDACTGTGAKLGSKMRVC 963
Query: 165 AGCQGSGMKISMRHLGANMIQQMQHPCNECKGTGETISDKDRCPQCKGEKVVQQK 219
+ C G G + + Q+ C C G GE IS+ C +C GE ++ K
Sbjct: 964 STCGGRGQVMRTEQTPFGLFSQVS-VCPNCGGDGEVISES--CRKCSGEGRIRVK 1119
>TC87379 similar to PIR|T04618|T04618 heat shock protein homolog F20O9.160 -
Arabidopsis thaliana, partial (75%)
Length = 1431
Score = 139 bits (349), Expect = 2e-33
Identities = 103/311 (33%), Positives = 142/311 (45%), Gaps = 18/311 (5%)
Frame = +2
Query: 14 YYEILGVSKNASPDDLKKAYKKAAIKNHPDKGGDPE-----KFKELAQAYEVLSDPEKRE 68
YYEIL V KNA+ D+LKKAY+K A+K HPDK D + KFK +++AYEVLSDP+KR
Sbjct: 95 YYEILEVDKNATDDELKKAYRKLAMKWHPDKNPDNKNDAETKFKLISEAYEVLSDPQKRA 274
Query: 69 VYDQYGEDALKEGMGGGGGGHDPFDIFSSFFGGGGFPGGGSSRGRRQRRGEDVVHPLKVS 128
+YDQYGE LK GM G P+ F R R R D +
Sbjct: 275 IYDQYGESNLKNGMPTAGDNAAPY-----------FQTHDGRRFRFNPRSADGIFAEVFG 421
Query: 129 LEDLYLGTSKKLSLSRNVLCSKCNGKG-------SKSGASMTCAGCQGSGMKISMRHLGA 181
Y G + C G G S+S + G + S
Sbjct: 422 FSSPYGGMGMR--------GGGCRGMGMRGQSWVSRSFGDIFGKDVFGESRQTSQAPRRK 577
Query: 182 NMIQQMQHPCNE---CKGTGETISDKDRCPQCKGEKVVQQKKVLEVHVEKGMQNGQKITF 238
+ + PC+ KGT + + G K V +++L + ++ G + G KITF
Sbjct: 578 APPIENKLPCSLEELYKGTTKKMKISREIAYASG-KTVPVEEILTIEIQPGWKKGTKITF 754
Query: 239 PGEADEAPDTVTGDIVFVLQQKEHPKFKRKGEDLFVEHTLSLTEALC---GFQFALTHLD 295
P + +E P+ + DIVFV+ +K H F R+G DL + + L E + F LT LD
Sbjct: 755 PEKGNEQPNVIAADIVFVIDEKPHNVFTRQGNDLVMTQKILLAEGEALSRSYTFQLTTLD 934
Query: 296 SRQLLIKSNPG 306
R L I + G
Sbjct: 935 GRGLTIAIDNG 967
>TC90074 similar to PIR|G84590|G84590 probable heat shock protein [imported]
- Arabidopsis thaliana, partial (88%)
Length = 1287
Score = 120 bits (302), Expect = 7e-28
Identities = 109/356 (30%), Positives = 150/356 (41%), Gaps = 53/356 (14%)
Frame = +1
Query: 14 YYEILGVSKNASPDDLKKAYKKAAIKNHPDKGGDPEK-----FKELAQAYEVLSDPEKRE 68
YY+IL V KNA+ ++LKKAY+K A+K HPDK +K FKE+++AYEVLSDP+K+
Sbjct: 13 YYKILKVDKNATEEELKKAYRKLAMKWHPDKNPSNKKDAEAKFKEISEAYEVLSDPQKKA 192
Query: 69 VYDQYGEDALKEGMG--------GGGGGHDPF--------DIFSSFF-----------GG 101
+YDQYGE+ LK + G G F DIF+ FF GG
Sbjct: 193 IYDQYGEEGLKGQVPPPQDATFFQSGDGPTTFRFNPRNANDIFAEFFGFSSPFGGMGAGG 372
Query: 102 GGFPGGGSSRG-------------------RRQ--RRGEDVVHPLKVSLEDLYLGTSKKL 140
G GG S G R+Q R+ + + L SLE+LY GT+KK+
Sbjct: 373 NGMRGGARSFGGMFGGDDHMFSSFDEGRPMRQQGPRKAAAIENRLPCSLEELYKGTTKKM 552
Query: 141 SLSRNVLCSKCNGKGSKSGASMTCAGCQGSGMKISMRHLGANMIQQMQHPCNECKGTGET 200
+SR + A G M + E
Sbjct: 553 KISREI------------------ADASGKTMPVE-----------------------EI 609
Query: 201 ISDKDRCPQCKGEKVVQQKKVLEVHVEKGMQNGQKITFPGEADEAPDTVTGDIVFVLQQK 260
++ + + KG K+ EKG N Q P + ++FV+ +K
Sbjct: 610 LTIEVKPGWKKGTKI--------TFPEKG--NEQPNVIPAD-----------LIFVIDEK 726
Query: 261 EHPKFKRKGEDLFVEHTLSLTEALCGFQFALTHLDSRQLLIKSNPGEVVKPGWMSV 316
H F R G DL +SL EAL G+ LT LD R L + N V+ P + V
Sbjct: 727 PHGVFTRDGNDLVATQKISLAEALTGYTVRLTTLDGRVLNVPIN--NVIHPSYEEV 888
>TC78759 similar to GP|21618097|gb|AAM67147.1 putative heat shock protein
{Arabidopsis thaliana}, partial (89%)
Length = 1344
Score = 106 bits (264), Expect = 2e-23
Identities = 72/179 (40%), Positives = 89/179 (49%), Gaps = 53/179 (29%)
Frame = +3
Query: 14 YYEILGVSKNASPDDLKKAYKKAAIKNHPDKGGDPEK-----FKELAQAYEVLSDPEKRE 68
YY++L + +NA DDLKKAY+K A+K HPDK + +K FK +++AY+VLSDP+KR
Sbjct: 81 YYKVLQIDRNAKDDDLKKAYRKLAMKWHPDKNPNNKKDAEAKFKTISEAYDVLSDPQKRA 260
Query: 69 VYDQYGEDALKEGM---------GGGGGGHDPF--------DIFSSFFG-----GG---- 102
VYDQYGE+ LK M GG GG F DIFS FFG GG
Sbjct: 261 VYDQYGEEGLKGQMPPPGAGGFSDGGDGGPTMFRFNPRSADDIFSEFFGFQRPFGGGMGD 440
Query: 103 --------GFPGG--------------GSSRGRRQRRGEDVVHPLKVSLEDLYLGTSKK 139
GFP G G R+ + L SLEDLY GT+KK
Sbjct: 441 MGGHPGASGFPRGMFRDDLFSSFRNSAGEGSANVMRKSAPIERTLPCSLEDLYKGTTKK 617
Score = 73.6 bits (179), Expect = 1e-13
Identities = 35/98 (35%), Positives = 56/98 (56%)
Frame = +1
Query: 219 KKVLEVHVEKGMQNGQKITFPGEADEAPDTVTGDIVFVLQQKEHPKFKRKGEDLFVEHTL 278
+++L + ++ G + G KITFP + +E + D+VF++ +K H FKR G DL V +
Sbjct: 670 EEILTIEIKPGWKKGTKITFPEKGNEQRGLIPADLVFIIDEKPHTVFKRDGNDLVVTQKI 849
Query: 279 SLTEALCGFQFALTHLDSRQLLIKSNPGEVVKPGWMSV 316
SL EAL G+ +T LD R L + N ++ P + V
Sbjct: 850 SLVEALTGYTAQITTLDGRNLTVPVN--TIISPSYEEV 957
>TC87441 similar to GP|21928031|gb|AAM78044.1 At3g62600/F26K9_30
{Arabidopsis thaliana}, partial (94%)
Length = 1563
Score = 103 bits (256), Expect = 1e-22
Identities = 70/222 (31%), Positives = 106/222 (47%), Gaps = 4/222 (1%)
Frame = +3
Query: 93 DIFSSFFGGGGFPGGGSSRGRRQRRGEDVVHPLKVSLEDLYLGTSKKLSLSRNVLCSKCN 152
DIF+SFFGGG + +G+DV+ L +LEDLY+G S K+ +NV+
Sbjct: 522 DIFNSFFGGGSM----EEEEEKIAKGDDVIVDLDATLEDLYMGGSLKVWREKNVV----- 674
Query: 153 GKGSKSGASMTCAGCQGSGMKISMRHLGANMIQQMQHPCNECKGTGETISDKDRCPQCKG 212
K C+ ++ R +G M QQM C QC
Sbjct: 675 ----KPAPGKRRCNCRN---EVYHRQIGPGMFQQMTEQV---------------CDQCAN 788
Query: 213 EKVVQQKKVLEVHVEKGMQNGQKITFPGEADEAPDTVTGDIVFVLQQKEHPKFKRKGEDL 272
K V++ + V +EKGM++GQ++ F + + D +GD+ F ++ H FKR+G DL
Sbjct: 789 VKYVREGYFVTVDIEKGMKDGQEVLFYEDGEPIIDGESGDLRFRIRTAPHELFKREGNDL 968
Query: 273 FVEHTLSLTEALCGFQFALTHLDSRQLLIKS----NPGEVVK 310
T++L +AL GF+ + HLD + I S NP +V K
Sbjct: 969 HTTVTITLVQALVGFEKTIKHLDEHLVDISSKGITNPKQVRK 1094
Score = 92.4 bits (228), Expect = 2e-19
Identities = 53/110 (48%), Positives = 67/110 (60%), Gaps = 6/110 (5%)
Frame = +2
Query: 14 YYEILGVSKNASPDDLKKAYKKAAIKNHPDKGGDPE----KFKELAQAYEVLSDPEKREV 69
YY+IL VSK AS D +K+AY+K A+K HPDK E KF E++ AYEVLSD EKR +
Sbjct: 266 YYDILQVSKGASDDQIKRAYRKLALKYHPDKNPGNEEANKKFAEISNAYEVLSDNEKRNI 445
Query: 70 YDQYGEDALKEGMGGG--GGGHDPFDIFSSFFGGGGFPGGGSSRGRRQRR 117
YD+YGE+ LK+ GG GGG++ F F G G +R RR
Sbjct: 446 YDKYGEEGLKQHAAGGGRGGGNEHAGYF*LFLWRGINGGRRRENCKR*RR 595
>BG647719 similar to GP|10177754|dbj DnaJ protein-like {Arabidopsis
thaliana}, partial (16%)
Length = 702
Score = 101 bits (251), Expect = 5e-22
Identities = 64/146 (43%), Positives = 82/146 (55%), Gaps = 4/146 (2%)
Frame = +3
Query: 14 YYEILGVSKNASPDDLKKAYKKAAIKNHPDKG-GDPE---KFKELAQAYEVLSDPEKREV 69
YY++LGVSK+AS ++KKAY A K HPD DPE KF+E+ AYEVL D EKR+
Sbjct: 261 YYDVLGVSKDASSSEIKKAYYGLAKKLHPDANKDDPEAEKKFQEVTLAYEVLKDGEKRQQ 440
Query: 70 YDQYGEDALKEGMGGGGGGHDPFDIFSSFFGGGGFPGGGSSRGRRQRRGEDVVHPLKVSL 129
YDQ G DA GG GG+ F+ F F GG SS ++ GEDV +++S
Sbjct: 441 YDQVGHDAYVNNQSGGFGGNGGFNPFEQMFRGG--HDIFSSFFHQKFGGEDVKTVVELSF 614
Query: 130 EDLYLGTSKKLSLSRNVLCSKCNGKG 155
+ G +K LS +VLC G G
Sbjct: 615 TEAIQGCTKTLSFQTDVLCMLVVGVG 692
>TC78760 similar to PIR|T48161|T48161 heat shock protein 40-like -
Arabidopsis thaliana, partial (56%)
Length = 762
Score = 96.7 bits (239), Expect = 1e-20
Identities = 58/126 (46%), Positives = 72/126 (57%), Gaps = 27/126 (21%)
Frame = +3
Query: 14 YYEILGVSKNASPDDLKKAYKKAAIKNHPDKGGDPEK-----FKELAQAYEVLSDPEKRE 68
YY++L + +NA DDLKKAY+K A+K HPDK + +K FK +++AY+VLSDP+KR
Sbjct: 27 YYKVLQIDRNAKDDDLKKAYRKLAMKWHPDKNPNNKKDAEAKFKTISEAYDVLSDPQKRA 206
Query: 69 VYDQYGEDALKEGM---------GGGGGGHDPF--------DIFSSFFG-----GGGFPG 106
VYDQYGE+ LK M GG GG F DIFS FFG GGG
Sbjct: 207 VYDQYGEEGLKGQMPPPGAGGFSDGGDGGPTMFRFNPRSADDIFSEFFGFQRPFGGGMGD 386
Query: 107 GGSSRG 112
G G
Sbjct: 387 MGGHPG 404
>TC93716 similar to GP|6179940|gb|AAF05720.1| DnaJ-like protein {Nicotiana
tabacum}, partial (33%)
Length = 571
Score = 93.6 bits (231), Expect = 1e-19
Identities = 56/127 (44%), Positives = 70/127 (55%), Gaps = 27/127 (21%)
Frame = +2
Query: 14 YYEILGVSKNASPDDLKKAYKKAAIKNHPDKGGDPEK-----FKELAQAYEVLSDPEKRE 68
YY++L V +NA DDLKKAY+K A+K HPDK + +K FK++++AY+VLSD KR
Sbjct: 143 YYKLLQVDRNAKDDDLKKAYRKLAMKWHPDKNPNNKKEAEAKFKQISEAYDVLSDSNKRA 322
Query: 69 VYDQYGEDALKEGM--------GGGGGGHD--------------PFDIFSSFFGGGGFPG 106
VYDQYGED LK M G GG + P DIF+ FFG G
Sbjct: 323 VYDQYGEDGLKGQMPPPPDSHASGSGGSRNFSPDDFPYQFNHRTPDDIFAEFFGDRNPFG 502
Query: 107 GGSSRGR 113
G GR
Sbjct: 503 GMGGMGR 523
>BG644408 homologue to PIR|T07371|T07 dnaJ protein homolog - potato, partial
(34%)
Length = 582
Score = 93.2 bits (230), Expect = 1e-19
Identities = 43/49 (87%), Positives = 46/49 (93%)
Frame = +1
Query: 263 PKFKRKGEDLFVEHTLSLTEALCGFQFALTHLDSRQLLIKSNPGEVVKP 311
PKFKRKG+DLFVEHTLSLTEALCGFQF LTHLD+RQL+IK PGEVVKP
Sbjct: 7 PKFKRKGDDLFVEHTLSLTEALCGFQFILTHLDNRQLIIKPQPGEVVKP 153
Score = 35.4 bits (80), Expect = 0.036
Identities = 17/34 (50%), Positives = 20/34 (58%)
Frame = +2
Query: 313 WMSVRRQHFMMSTLKKRREEGSKLSKRHMMRMMK 346
W S RR +MM T K+R S KRHM +MMK
Sbjct: 332 WTSARRPPYMMLTSKRRCGGSSNRPKRHMTKMMK 433
>AJ503880 similar to GP|12838381|dbj DnaJ (Hsp40) homolog subfamily B
member 6~data source:MGD source key:MGI:1344381
evidence:ISS~, partial (26%)
Length = 466
Score = 92.8 bits (229), Expect = 2e-19
Identities = 53/117 (45%), Positives = 71/117 (60%), Gaps = 16/117 (13%)
Frame = +1
Query: 14 YYEILGVSKNASPDDLKKAYKKAAIKNHPDKGGDPE-----KFKELAQAYEVLSDPEKRE 68
YYEILG+ +AS D+++KAY+K A+ HPDK + KFK++A+AYEVLSD +KR+
Sbjct: 43 YYEILGIEVDASVDEIRKAYRKQALLWHPDKNVQRKEEAEAKFKKIAEAYEVLSDVDKRQ 222
Query: 69 VYDQYGEDALKEGMGG---------GGGGHDPFDIFSSFFGG--GGFPGGGSSRGRR 114
+Y++YGE+ LK G HDP +IF SFFGG GGG G R
Sbjct: 223 IYNKYGEEGLKNGEPNEYERNNYTPNFHFHDPEEIFRSFFGGSLAEIFGGGGLGGMR 393
>TC77858 similar to GP|20197886|gb|AAD22362.2 putative DnaJ protein
{Arabidopsis thaliana}, partial (78%)
Length = 1869
Score = 92.4 bits (228), Expect = 2e-19
Identities = 48/85 (56%), Positives = 59/85 (68%), Gaps = 6/85 (7%)
Frame = +3
Query: 14 YYEILGVSKNASPDDLKKAYKKAAIKNHPDKGGDP---EKFKELAQAYEVLSDPEKREVY 70
YY +LGVSKN++ ++K AY+K A HPD DP EKFKE++ AYEVLSD EKR +Y
Sbjct: 501 YYTVLGVSKNSTKSEIKTAYRKLARNYHPDVNKDPGAEEKFKEISNAYEVLSDDEKRSIY 680
Query: 71 DQYGEDALK--EGMGGG-GGGHDPF 92
D+YGE LK +G GGG GG PF
Sbjct: 681 DKYGEAGLKGSQGFGGGMGGFQQPF 755
Score = 87.4 bits (215), Expect = 8e-18
Identities = 57/216 (26%), Positives = 95/216 (43%), Gaps = 2/216 (0%)
Frame = +1
Query: 82 MGGGGGGHDPFDIFSSFFGGGGFPGGGSSRGRRQRRGEDVVHPLKVSLEDLYLGTSKKLS 141
+G G +PFD+F S F G G GED + L ++ ++ G K++
Sbjct: 724 VGAWGDFSNPFDLFESLFEG--MDGRSRQSWDGAMNGEDEYYSLVLNFKEAVFGVEKEIE 897
Query: 142 LSRNVLCSKCNGKGSKSGA-SMTCAGCQGSGMKISMRHLGANMIQQMQHPCNECKGTGET 200
+ R C C+G G+K G S C C G G ++ + QQ C+ C GTGET
Sbjct: 898 IRRLEKCGTCDGSGAKPGTKSSRCNTCGGQGRVVTQTRTPLGIFQQSM-TCSSCNGTGET 1074
Query: 201 ISDKDRCPQCKGEKVVQQKKVLEVHVEKGMQNGQKITFPGEADEAP-DTVTGDIVFVLQQ 259
+ C C G+ V++ K + + V G+ +G ++ E + GD+ VL+
Sbjct: 1075---RTPCSTCSGDGRVRKTKRISLKVPAGVDSGSRLRVRNEGNSGKRGGSPGDLFVVLEV 1245
Query: 260 KEHPKFKRKGEDLFVEHTLSLTEALCGFQFALTHLD 295
P KR+ ++ +S +A+ G + +D
Sbjct: 1246IPDPVLKREDTNILYSCKVSYIDAILGTTIKVPTVD 1353
>TC81458 similar to GP|21553367|gb|AAM62460.1 DnaJ protein-like {Arabidopsis
thaliana}, partial (38%)
Length = 1178
Score = 89.7 bits (221), Expect = 2e-18
Identities = 57/192 (29%), Positives = 102/192 (52%), Gaps = 3/192 (1%)
Frame = +2
Query: 118 GEDVVHPLKVSLEDLYLGTSKKLSLSRNVLCSKCNGKGSKSGASM-TCAGCQGSGMKISM 176
G+DV +++S + G +K ++ +++C+ C G G G TC C+GSG ++
Sbjct: 23 GQDVKTFVELSFMEAVRGCAKTITFQTDMICNTCGGSGVPPGTRPETCKRCKGSG--VTS 196
Query: 177 RHLGANMIQQMQHPCNECKGTGETISDKDRCPQCKGEKVVQQKKVLEVHVEKGMQNGQ-- 234
G I +M+ C C+G+G+ + K C CKG KV + K +++ + G+ N +
Sbjct: 197 VQAG---IFRMETTCGACRGSGKIV--KSFCKSCKGAKVNRATKSVKLDIMAGIDNNETI 361
Query: 235 KITFPGEADEAPDTVTGDIVFVLQQKEHPKFKRKGEDLFVEHTLSLTEALCGFQFALTHL 294
K+ G AD D GD+ ++ +E P F+R+G D+ V+ LS+T+A+ G + L
Sbjct: 362 KVYRSGGADPEGDR-PGDLYVTIKVREDPVFRREGSDIHVDTVLSITQAILGGTIQVPTL 538
Query: 295 DSRQLLIKSNPG 306
+ +++K PG
Sbjct: 539 -TGDVVLKVRPG 571
>TC85119 similar to PIR|G84590|G84590 probable heat shock protein [imported]
- Arabidopsis thaliana, partial (72%)
Length = 796
Score = 77.4 bits (189), Expect = 8e-15
Identities = 39/103 (37%), Positives = 59/103 (56%)
Frame = +3
Query: 214 KVVQQKKVLEVHVEKGMQNGQKITFPGEADEAPDTVTGDIVFVLQQKEHPKFKRKGEDLF 273
K + +++L + V+ G + G KITFP + +E P+ D++FV+ ++ H F R+G DL
Sbjct: 336 KTMPVEEILTITVKPGWKKGTKITFPEKGNEQPNVTAADLIFVIDERPHSVFSREGNDLI 515
Query: 274 VEHTLSLTEALCGFQFALTHLDSRQLLIKSNPGEVVKPGWMSV 316
V +SL EAL G+ LT LD R L I N V+ P + V
Sbjct: 516 VTQKISLAEALTGYTVHLTTLDGRNLSIPIN--NVIHPNYEEV 638
Score = 30.0 bits (66), Expect = 1.5
Identities = 22/86 (25%), Positives = 34/86 (38%), Gaps = 32/86 (37%)
Frame = +3
Query: 93 DIFSSFF-------GGGGFPGGGSSRGR-------------------------RQRRGED 120
DIF+ FF G GG GGG R R R+
Sbjct: 63 DIFAEFFGFSSPFGGMGGRGGGGGMRSRFSGGMFGDDMFGSFGEGGGIHMSQAAPRKAPA 242
Query: 121 VVHPLKVSLEDLYLGTSKKLSLSRNV 146
+ + L +L+++Y GT+K + +SR +
Sbjct: 243 IDNKLSCTLDEIYRGTTKNMKISREI 320
>TC82757 similar to PIR|T08563|T08563 dnaJ-related protein T22F8.50 -
Arabidopsis thaliana, partial (95%)
Length = 1317
Score = 74.7 bits (182), Expect = 5e-14
Identities = 46/102 (45%), Positives = 57/102 (55%), Gaps = 4/102 (3%)
Frame = +2
Query: 7 KKSDNTKYYEILGVSKNASPDDLKKAYKKAAIKNHPDKG-GDP---EKFKELAQAYEVLS 62
K T YY+ LGVS +AS D+KKAY A HPDK GDP E F+ L +AY+VLS
Sbjct: 92 KMVKETAYYDTLGVSVDASAADIKKAYYVKARIVHPDKNPGDPKAAENFQLLGEAYQVLS 271
Query: 63 DPEKREVYDQYGEDALKEGMGGGGGGHDPFDIFSSFFGGGGF 104
DPEKRE YD+ G+ + + DP +F FG F
Sbjct: 272 DPEKREAYDKNGKAGVSQ-----DAMMDPTTVFGMLFGSEFF 382
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.316 0.136 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 11,106,632
Number of Sequences: 36976
Number of extensions: 155733
Number of successful extensions: 2475
Number of sequences better than 10.0: 171
Number of HSP's better than 10.0 without gapping: 1436
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1939
length of query: 375
length of database: 9,014,727
effective HSP length: 98
effective length of query: 277
effective length of database: 5,391,079
effective search space: 1493328883
effective search space used: 1493328883
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)
S2: 59 (27.3 bits)
Medicago: description of AC133862.8


