

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC133709.13 - phase: 0
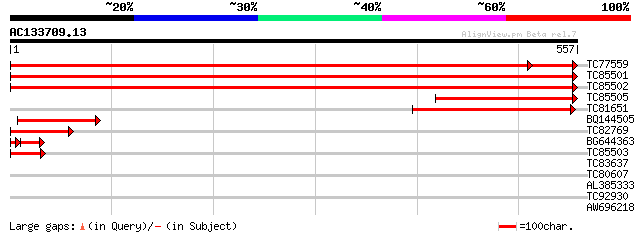
(557 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC77559 homologue to SP|Q9SMK9|PAL2_CICAR Phenylalanine ammonia-... 1010 0.0
TC85501 homologue to SP|P45732|PALY_STYHU Phenylalanine ammonia-... 927 0.0
TC85502 homologue to SP|P27990|PALY_MEDSA Phenylalanine ammonia-... 920 0.0
TC85505 homologue to SP|P27990|PALY_MEDSA Phenylalanine ammonia-... 218 6e-57
TC81651 similar to SP|P19143|PAL3_PHAVU Phenylalanine ammonia-ly... 201 6e-52
BQ144505 similar to SP|P45734|PALY_ Phenylalanine ammonia-lyase ... 105 3e-23
TC82769 similar to SP|P19143|PAL3_PHAVU Phenylalanine ammonia-ly... 103 2e-22
BG644363 homologue to SP|P26600|PAL5 Phenylalanine ammonia-lyase... 45 9e-06
TC85503 homologue to SP|P27990|PALY_MEDSA Phenylalanine ammonia-... 48 1e-05
TC83637 similar to GP|9279731|dbj|BAB01321.1 disease resistance ... 33 0.22
TC80607 weakly similar to PIR|D86431|D86431 protein T5I8.6 [impo... 29 5.4
AL385333 28 7.1
TC92930 similar to GP|13430332|gb|AAK25798.1 rubisco activase {Z... 28 9.3
AW696218 homologue to PIR|B96835|B96 CAD ATPase (AAA1) 35570-33... 28 9.3
>TC77559 homologue to SP|Q9SMK9|PAL2_CICAR Phenylalanine ammonia-lyase 2 (EC
4.3.1.5). [Chickpea Garbanzo] {Cicer arietinum}, partial
(96%)
Length = 2406
Score = 1010 bits (2612), Expect(2) = 0.0
Identities = 513/513 (100%), Positives = 513/513 (100%)
Frame = +3
Query: 1 MLVRINTLLQGYSGIRFEIMEAIAKFLNHNITPCLPLRGTITASGDLIPLSYVAGLLIGR 60
MLVRINTLLQGYSGIRFEIMEAIAKFLNHNITPCLPLRGTITASGDLIPLSYVAGLLIGR
Sbjct: 564 MLVRINTLLQGYSGIRFEIMEAIAKFLNHNITPCLPLRGTITASGDLIPLSYVAGLLIGR 743
Query: 61 PNSKSIGPNGQVLNAQEAFQLAGIETGFFELQPKEGLALVNGTAVGSGLASLVLFDTNLL 120
PNSKSIGPNGQVLNAQEAFQLAGIETGFFELQPKEGLALVNGTAVGSGLASLVLFDTNLL
Sbjct: 744 PNSKSIGPNGQVLNAQEAFQLAGIETGFFELQPKEGLALVNGTAVGSGLASLVLFDTNLL 923
Query: 121 VVLSEILSAIFAEVMLGKPQFTDHLIHKVKLHPGQIEAAAIMEHILDGSYYGKAAQKVHE 180
VVLSEILSAIFAEVMLGKPQFTDHLIHKVKLHPGQIEAAAIMEHILDGSYYGKAAQKVHE
Sbjct: 924 VVLSEILSAIFAEVMLGKPQFTDHLIHKVKLHPGQIEAAAIMEHILDGSYYGKAAQKVHE 1103
Query: 181 INPLQKPKQDRYAIRTSPQWLGPQIEVIRYATKMIEREINSVNDNPLIDVSRDKALHGGN 240
INPLQKPKQDRYAIRTSPQWLGPQIEVIRYATKMIEREINSVNDNPLIDVSRDKALHGGN
Sbjct: 1104 INPLQKPKQDRYAIRTSPQWLGPQIEVIRYATKMIEREINSVNDNPLIDVSRDKALHGGN 1283
Query: 241 FQGTPIGVSMDNTRLAIAAIGKLMFAQFTELVNDIYNNGLPSSLTASRNPSLDYGFKGAE 300
FQGTPIGVSMDNTRLAIAAIGKLMFAQFTELVNDIYNNGLPSSLTASRNPSLDYGFKGAE
Sbjct: 1284 FQGTPIGVSMDNTRLAIAAIGKLMFAQFTELVNDIYNNGLPSSLTASRNPSLDYGFKGAE 1463
Query: 301 VAMASYCSELQYLASPVTTHVQSAEQHNQDVNSLGLISARKTAEAVEIWKLMSSTFLVAL 360
VAMASYCSELQYLASPVTTHVQSAEQHNQDVNSLGLISARKTAEAVEIWKLMSSTFLVAL
Sbjct: 1464 VAMASYCSELQYLASPVTTHVQSAEQHNQDVNSLGLISARKTAEAVEIWKLMSSTFLVAL 1643
Query: 361 CQAIDLRHIEENFKSVVKNTVSQVAKRILTVGVNGELHPSRFCEKDLLNVVEGEYVFTYI 420
CQAIDLRHIEENFKSVVKNTVSQVAKRILTVGVNGELHPSRFCEKDLLNVVEGEYVFTYI
Sbjct: 1644 CQAIDLRHIEENFKSVVKNTVSQVAKRILTVGVNGELHPSRFCEKDLLNVVEGEYVFTYI 1823
Query: 421 DDPCSPIYPLMQKLRYVLVDHALQNGDKEANSSTSIFQKIGAFEEELKALLPKEVENARV 480
DDPCSPIYPLMQKLRYVLVDHALQNGDKEANSSTSIFQKIGAFEEELKALLPKEVENARV
Sbjct: 1824 DDPCSPIYPLMQKLRYVLVDHALQNGDKEANSSTSIFQKIGAFEEELKALLPKEVENARV 2003
Query: 481 EIENGNPAVPNMIKECRSYPLYKFMRETLGTSL 513
EIENGNPAVPNMIKECRSYPLYKFMRETLGTSL
Sbjct: 2004 EIENGNPAVPNMIKECRSYPLYKFMRETLGTSL 2102
Score = 100 bits (250), Expect(2) = 0.0
Identities = 44/48 (91%), Positives = 45/48 (93%)
Frame = +1
Query: 510 GTSLLTGEKIRSPGEDCDKVFTAMCDGRFIDPMLDCLKEWNGVPLPIC 557
G LTGEKI+SPGEDCDKVFTAMCDGRFIDPMLDCLKEWNGVPLPIC
Sbjct: 2092 GQVCLTGEKIKSPGEDCDKVFTAMCDGRFIDPMLDCLKEWNGVPLPIC 2235
>TC85501 homologue to SP|P45732|PALY_STYHU Phenylalanine ammonia-lyase (EC
4.3.1.5). [Townsville stylo] {Stylosanthes humilis},
partial (96%)
Length = 2567
Score = 927 bits (2397), Expect = 0.0
Identities = 451/557 (80%), Positives = 507/557 (90%)
Frame = +3
Query: 1 MLVRINTLLQGYSGIRFEIMEAIAKFLNHNITPCLPLRGTITASGDLIPLSYVAGLLIGR 60
MLVRINTLLQGYSGIRFEI+EAI K LN+NITPCLPLRGTITASGDL+PLSY+AGLL GR
Sbjct: 597 MLVRINTLLQGYSGIRFEILEAITKLLNNNITPCLPLRGTITASGDLVPLSYIAGLLTGR 776
Query: 61 PNSKSIGPNGQVLNAQEAFQLAGIETGFFELQPKEGLALVNGTAVGSGLASLVLFDTNLL 120
PNSK++GP+G++LNA+EAFQLAGI + FFELQPKEGLALVNGTAVGSGLAS+VLF+ N+L
Sbjct: 777 PNSKAVGPSGEILNAKEAFQLAGIGSDFFELQPKEGLALVNGTAVGSGLASIVLFEANVL 956
Query: 121 VVLSEILSAIFAEVMLGKPQFTDHLIHKVKLHPGQIEAAAIMEHILDGSYYGKAAQKVHE 180
VLSE++SAIFAEVM GKP+FTDHL HK+K HPGQIEAAAIMEHILDGS Y KAA+K+HE
Sbjct: 957 AVLSEVMSAIFAEVMQGKPEFTDHLTHKLKHHPGQIEAAAIMEHILDGSAYVKAAKKLHE 1136
Query: 181 INPLQKPKQDRYAIRTSPQWLGPQIEVIRYATKMIEREINSVNDNPLIDVSRDKALHGGN 240
+PLQKPKQDRYA+RTSPQWLGP IEVIR++TK IEREINSVNDNPLIDVSR+KA+HGGN
Sbjct: 1137 TDPLQKPKQDRYALRTSPQWLGPLIEVIRFSTKSIEREINSVNDNPLIDVSRNKAIHGGN 1316
Query: 241 FQGTPIGVSMDNTRLAIAAIGKLMFAQFTELVNDIYNNGLPSSLTASRNPSLDYGFKGAE 300
FQGTPIGVSMDNTRLA+A+IGKLMFAQF+ELVND YNNGLPS+LTASRNPSLDYGFKG+E
Sbjct: 1317 FQGTPIGVSMDNTRLALASIGKLMFAQFSELVNDFYNNGLPSNLTASRNPSLDYGFKGSE 1496
Query: 301 VAMASYCSELQYLASPVTTHVQSAEQHNQDVNSLGLISARKTAEAVEIWKLMSSTFLVAL 360
+AMASYCSELQYLA+PVTTHVQSAEQHNQDVNSLGLIS+RKT EA+EI KLMSSTFL+AL
Sbjct: 1497 IAMASYCSELQYLANPVTTHVQSAEQHNQDVNSLGLISSRKTNEAIEILKLMSSTFLIAL 1676
Query: 361 CQAIDLRHIEENFKSVVKNTVSQVAKRILTVGVNGELHPSRFCEKDLLNVVEGEYVFTYI 420
CQAIDLRH+EEN ++ VKNTVSQVAKR LT GVNGELHPSRFCEKDLL VV+ EYVF Y
Sbjct: 1677 CQAIDLRHLEENLRNTVKNTVSQVAKRTLTTGVNGELHPSRFCEKDLLKVVDREYVFAYA 1856
Query: 421 DDPCSPIYPLMQKLRYVLVDHALQNGDKEANSSTSIFQKIGAFEEELKALLPKEVENARV 480
DDPC YPLMQKLR VLVDHAL N + E NS+TSIFQKI FE+ELKA+LPKEVE+AR
Sbjct: 1857 DDPCLATYPLMQKLRQVLVDHALVNTEGEKNSNTSIFQKIATFEDELKAILPKEVESART 2036
Query: 481 EIENGNPAVPNMIKECRSYPLYKFMRETLGTSLLTGEKIRSPGEDCDKVFTAMCDGRFID 540
ENG + N IKECRSYPLYKF+RE LGT+LLTGEK+ SPGE+CDK+FTAMC G+ +D
Sbjct: 2037 AYENGQSGISNKIKECRSYPLYKFVREELGTALLTGEKVISPGEECDKLFTAMCQGKIVD 2216
Query: 541 PMLDCLKEWNGVPLPIC 557
P+++CL EWNG PLPIC
Sbjct: 2217 PLMECLGEWNGAPLPIC 2267
>TC85502 homologue to SP|P27990|PALY_MEDSA Phenylalanine ammonia-lyase (EC
4.3.1.5). [Alfalfa] {Medicago sativa}, complete
Length = 2415
Score = 920 bits (2378), Expect = 0.0
Identities = 449/557 (80%), Positives = 503/557 (89%)
Frame = +2
Query: 1 MLVRINTLLQGYSGIRFEIMEAIAKFLNHNITPCLPLRGTITASGDLIPLSYVAGLLIGR 60
MLVRINTLLQGYSGIRFEI+EAI LN+N+TPCLPLRGTITASGDL+PLSY+AGLL GR
Sbjct: 608 MLVRINTLLQGYSGIRFEILEAITNLLNNNVTPCLPLRGTITASGDLVPLSYIAGLLTGR 787
Query: 61 PNSKSIGPNGQVLNAQEAFQLAGIETGFFELQPKEGLALVNGTAVGSGLASLVLFDTNLL 120
PNSK+ GP+G++LNA+EAF LAGI FFELQPKEGLALVNGTAVGSGLAS+VLF+ N+L
Sbjct: 788 PNSKAHGPSGEILNAKEAFALAGINAEFFELQPKEGLALVNGTAVGSGLASIVLFEANIL 967
Query: 121 VVLSEILSAIFAEVMLGKPQFTDHLIHKVKLHPGQIEAAAIMEHILDGSYYGKAAQKVHE 180
VLSE+LSAIFAEVM GKP+FTDHL HK+K HPGQIEAAAIMEHILDGS Y KA +K+HE
Sbjct: 968 AVLSEVLSAIFAEVMQGKPEFTDHLTHKLKHHPGQIEAAAIMEHILDGSAYVKADKKLHE 1147
Query: 181 INPLQKPKQDRYAIRTSPQWLGPQIEVIRYATKMIEREINSVNDNPLIDVSRDKALHGGN 240
++PLQKPKQDRYA+RTSPQWLGP +EVIR++TK IEREINSVNDNPLIDVSR+KALHGGN
Sbjct: 1148 MDPLQKPKQDRYALRTSPQWLGPLVEVIRFSTKSIEREINSVNDNPLIDVSRNKALHGGN 1327
Query: 241 FQGTPIGVSMDNTRLAIAAIGKLMFAQFTELVNDIYNNGLPSSLTASRNPSLDYGFKGAE 300
FQGTPIGVSMDNTRLA+A+IGKLMFAQF+ELVND YNNGLPS+L+ASRNPSLDYGFKGAE
Sbjct: 1328 FQGTPIGVSMDNTRLALASIGKLMFAQFSELVNDFYNNGLPSNLSASRNPSLDYGFKGAE 1507
Query: 301 VAMASYCSELQYLASPVTTHVQSAEQHNQDVNSLGLISARKTAEAVEIWKLMSSTFLVAL 360
+AMASYCSELQYLA+PVTTHVQSAEQHNQDVNSLGLIS+RKT EA+EI +LMSSTFL+AL
Sbjct: 1508 IAMASYCSELQYLANPVTTHVQSAEQHNQDVNSLGLISSRKTYEAIEILQLMSSTFLIAL 1687
Query: 361 CQAIDLRHIEENFKSVVKNTVSQVAKRILTVGVNGELHPSRFCEKDLLNVVEGEYVFTYI 420
CQAIDLRH+EEN K+ VKNTVSQVAK+ LT+GVNGELHPSRFCEKDLL VV+ E+VF YI
Sbjct: 1688 CQAIDLRHLEENLKNSVKNTVSQVAKKTLTIGVNGELHPSRFCEKDLLKVVDREHVFAYI 1867
Query: 421 DDPCSPIYPLMQKLRYVLVDHALQNGDKEANSSTSIFQKIGAFEEELKALLPKEVENARV 480
DDPCS YPL QKLR VLVDHAL NG+ E N +TSIFQKI FEEELK+LLPKEVE+AR
Sbjct: 1868 DDPCSATYPLSQKLRQVLVDHALVNGESEKNLNTSIFQKIATFEEELKSLLPKEVESART 2047
Query: 481 EIENGNPAVPNMIKECRSYPLYKFMRETLGTSLLTGEKIRSPGEDCDKVFTAMCDGRFID 540
E+GNP +PN I CRSYPLYKF+RE LGT LLTGE + SPGE CDK+FTAMC G+ ID
Sbjct: 2048 AYESGNPTIPNKINGCRSYPLYKFVREELGTGLLTGENVISPGEVCDKLFTAMCQGKIID 2227
Query: 541 PMLDCLKEWNGVPLPIC 557
P+L+CL EWNG PLPIC
Sbjct: 2228 PLLECLGEWNGAPLPIC 2278
>TC85505 homologue to SP|P27990|PALY_MEDSA Phenylalanine ammonia-lyase (EC
4.3.1.5). [Alfalfa] {Medicago sativa}, partial (19%)
Length = 652
Score = 218 bits (554), Expect = 6e-57
Identities = 101/139 (72%), Positives = 113/139 (80%)
Frame = +1
Query: 419 YIDDPCSPIYPLMQKLRYVLVDHALQNGDKEANSSTSIFQKIGAFEEELKALLPKEVENA 478
YIDDPCS YPL QKLR VLVDHAL NG+ E N +TSIFQKI FEEELK+LLPKEVE+A
Sbjct: 1 YIDDPCSATYPLSQKLRQVLVDHALVNGESEKNLNTSIFQKIATFEEELKSLLPKEVESA 180
Query: 479 RVEIENGNPAVPNMIKECRSYPLYKFMRETLGTSLLTGEKIRSPGEDCDKVFTAMCDGRF 538
R E+GNP +PN I CRSYPLYKF+RE LGT LLTGE + SPGE CDK+FTAMC G+
Sbjct: 181 RTAYESGNPTIPNKINGCRSYPLYKFVREELGTGLLTGENVISPGEVCDKLFTAMCQGKI 360
Query: 539 IDPMLDCLKEWNGVPLPIC 557
IDP+L+CL EWNG PLPIC
Sbjct: 361 IDPLLECLGEWNGAPLPIC 417
>TC81651 similar to SP|P19143|PAL3_PHAVU Phenylalanine ammonia-lyase class
III (EC 4.3.1.5). [Kidney bean French bean] {Phaseolus
vulgaris}, partial (23%)
Length = 708
Score = 201 bits (511), Expect = 6e-52
Identities = 95/161 (59%), Positives = 122/161 (75%)
Frame = +2
Query: 396 ELHPSRFCEKDLLNVVEGEYVFTYIDDPCSPIYPLMQKLRYVLVDHALQNGDKEANSSTS 455
E+ P R CE++LL VV+ EYVF+YIDDP + YPLM KL+ VL +HA + ++ +S
Sbjct: 26 EIDPFRHCEENLLKVVDREYVFSYIDDPFNVTYPLMPKLKQVLYEHAHISAINNKDAKSS 205
Query: 456 IFQKIGAFEEELKALLPKEVENARVEIENGNPAVPNMIKECRSYPLYKFMRETLGTSLLT 515
F+KIGAFE+ELK+LLPKEVE+ARV E GN +PN IKECRSYPLYKF+RE L LLT
Sbjct: 206 TFEKIGAFEDELKSLLPKEVESARVAFEKGNSEIPNRIKECRSYPLYKFVREELKIGLLT 385
Query: 516 GEKIRSPGEDCDKVFTAMCDGRFIDPMLDCLKEWNGVPLPI 556
GEK +P E+ +KVFTAMC + +DP+L+CL +W GVP+PI
Sbjct: 386 GEKDVTPDEEFEKVFTAMCQAKIVDPILECLGDWKGVPIPI 508
>BQ144505 similar to SP|P45734|PALY_ Phenylalanine ammonia-lyase (EC
4.3.1.5). [Subterranean clover] {Trifolium
subterraneum}, partial (11%)
Length = 863
Score = 105 bits (263), Expect = 3e-23
Identities = 52/82 (63%), Positives = 60/82 (72%)
Frame = +2
Query: 8 LLQGYSGIRFEIMEAIAKFLNHNITPCLPLRGTITASGDLIPLSYVAGLLIGRPNSKSIG 67
LLQGYSGI FEI+EAI LN+N+TPC PL GTITAS D +PLS++AGLL G PN K G
Sbjct: 11 LLQGYSGITFEILEAITNLLNNNVTPCSPLAGTITASRDSVPLSHIAGLLTGIPNCKPHG 190
Query: 68 PNGQVLNAQEAFQLAGIETGFF 89
P GQ+LNA+ F L GI F
Sbjct: 191PCGQILNAKHPFALTGINAESF 256
>TC82769 similar to SP|P19143|PAL3_PHAVU Phenylalanine ammonia-lyase class
III (EC 4.3.1.5). [Kidney bean French bean] {Phaseolus
vulgaris}, partial (27%)
Length = 652
Score = 103 bits (256), Expect = 2e-22
Identities = 49/62 (79%), Positives = 54/62 (87%)
Frame = +2
Query: 1 MLVRINTLLQGYSGIRFEIMEAIAKFLNHNITPCLPLRGTITASGDLIPLSYVAGLLIGR 60
MLVR+NTLLQGYSGIRFEI+ AI K LNHN+TP LPLRGT+TASGDLIPLSY+A LL G
Sbjct: 467 MLVRVNTLLQGYSGIRFEILXAITKLLNHNVTPILPLRGTVTASGDLIPLSYIAALLTGX 646
Query: 61 PN 62
N
Sbjct: 647 RN 652
>BG644363 homologue to SP|P26600|PAL5 Phenylalanine ammonia-lyase (EC
4.3.1.5) (PAL). [Tomato] {Lycopersicon esculentum},
partial (26%)
Length = 687
Score = 45.1 bits (105), Expect(2) = 9e-06
Identities = 19/24 (79%), Positives = 21/24 (87%)
Frame = +2
Query: 11 GYSGIRFEIMEAIAKFLNHNITPC 34
GYSGIRFEI+EAI K +N NITPC
Sbjct: 614 GYSGIRFEILEAITKLINSNITPC 685
Score = 22.3 bits (46), Expect(2) = 9e-06
Identities = 10/10 (100%), Positives = 10/10 (100%)
Frame = +3
Query: 1 MLVRINTLLQ 10
MLVRINTLLQ
Sbjct: 585 MLVRINTLLQ 614
>TC85503 homologue to SP|P27990|PALY_MEDSA Phenylalanine ammonia-lyase (EC
4.3.1.5). [Alfalfa] {Medicago sativa}, partial (27%)
Length = 724
Score = 47.8 bits (112), Expect = 1e-05
Identities = 24/35 (68%), Positives = 25/35 (70%)
Frame = +3
Query: 1 MLVRINTLLQGYSGIRFEIMEAIAKFLNHNITPCL 35
MLVRINTLLQGYSGI FEI+EAI L CL
Sbjct: 618 MLVRINTLLQGYSGIXFEILEAITNLLTTTSLHCL 722
>TC83637 similar to GP|9279731|dbj|BAB01321.1 disease resistance protein
RPP1-WsB {Arabidopsis thaliana}, partial (2%)
Length = 1045
Score = 33.5 bits (75), Expect = 0.22
Identities = 21/87 (24%), Positives = 44/87 (50%), Gaps = 1/87 (1%)
Frame = +3
Query: 336 LISARKTAEAVEIWKLMSSTFLVALCQAIDLRHIEENFKSV-VKNTVSQVAKRILTVGVN 394
L++ + + +VEI + + L + +EN K V N ++ + ++ +G+N
Sbjct: 579 LLALPELSSSVEILLVHCDSLKSVLFPSTIAEQFKENKKEVKFWNCLNLDERSLINIGLN 758
Query: 395 GELHPSRFCEKDLLNVVEGEYVFTYID 421
+++ +F +DL V +YV TY+D
Sbjct: 759 LQINLMKFAYQDLSTVEHDDYVETYVD 839
>TC80607 weakly similar to PIR|D86431|D86431 protein T5I8.6 [imported] -
Arabidopsis thaliana, partial (17%)
Length = 907
Score = 28.9 bits (63), Expect = 5.4
Identities = 19/69 (27%), Positives = 30/69 (42%)
Frame = +1
Query: 381 VSQVAKRILTVGVNGELHPSRFCEKDLLNVVEGEYVFTYIDDPCSPIYPLMQKLRYVLVD 440
+ +VA + L V G + S D N+ +G Y+F + D C+P + VD
Sbjct: 40 ICKVANKPLVVTYTGLMQASL----DSGNIQDGSYIFEKMKDICAPNLVTYNIMLKAYVD 207
Query: 441 HALQNGDKE 449
H + KE
Sbjct: 208 HGMFREAKE 234
>AL385333
Length = 310
Score = 28.5 bits (62), Expect = 7.1
Identities = 12/38 (31%), Positives = 22/38 (57%)
Frame = +2
Query: 327 HNQDVNSLGLISARKTAEAVEIWKLMSSTFLVALCQAI 364
H +NSL L+ R T+ + +WK+ S L+ + +A+
Sbjct: 62 HGMKMNSLSLMLERLTSMKIFLWKIHKSLSLILILKAM 175
>TC92930 similar to GP|13430332|gb|AAK25798.1 rubisco activase {Zantedeschia
aethiopica}, partial (42%)
Length = 765
Score = 28.1 bits (61), Expect = 9.3
Identities = 15/38 (39%), Positives = 24/38 (62%), Gaps = 4/38 (10%)
Frame = +2
Query: 146 IHKVKLHPGQIEAAAI----MEHILDGSYYGKAAQKVH 179
+ +V+L +E AA+ + I GS+YGKAAQ+V+
Sbjct: 356 VKRVQLADKYLEGAALGDANQDAIKSGSFYGKAAQQVN 469
>AW696218 homologue to PIR|B96835|B96 CAD ATPase (AAA1) 35570-33019
[imported] - Arabidopsis thaliana, partial (34%)
Length = 556
Score = 28.1 bits (61), Expect = 9.3
Identities = 25/84 (29%), Positives = 35/84 (40%), Gaps = 8/84 (9%)
Frame = -1
Query: 417 FTYIDDPCSPIYPL------MQKLRYVLVDHALQNGDKE--ANSSTSIFQKIGAFEEELK 468
FT +DD CSP PL + ++ VL AL +K S + + EE LK
Sbjct: 415 FTLLDDSCSPEAPLALHRESISSMKIVLGA*ALARSNKHRTIRSLSPRHLEAKVAEETLK 236
Query: 469 ALLPKEVENARVEIENGNPAVPNM 492
++P V A P PN+
Sbjct: 235 NVVPHSVATALARSVFPVPGGPNI 164
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.319 0.136 0.395
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 14,979,992
Number of Sequences: 36976
Number of extensions: 185517
Number of successful extensions: 880
Number of sequences better than 10.0: 28
Number of HSP's better than 10.0 without gapping: 880
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 880
length of query: 557
length of database: 9,014,727
effective HSP length: 101
effective length of query: 456
effective length of database: 5,280,151
effective search space: 2407748856
effective search space used: 2407748856
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 61 (28.1 bits)
Medicago: description of AC133709.13


