

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC131239.17 + phase: 0 /pseudo
(892 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
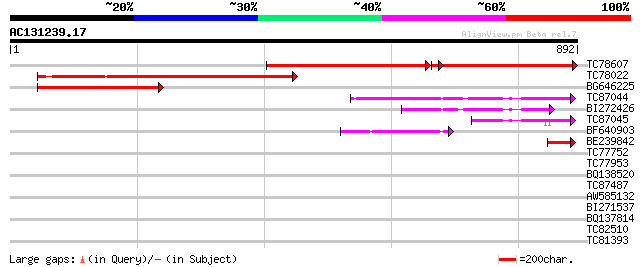
Score E
Sequences producing significant alignments: (bits) Value
TC78607 similar to GP|20466746|gb|AAM20690.1 putative protein {A... 516 0.0
TC78022 similar to GP|20466746|gb|AAM20690.1 putative protein {A... 496 e-140
BG646225 similar to GP|20466746|gb putative protein {Arabidopsis... 394 e-110
TC87044 similar to GP|21554145|gb|AAM63225.1 unknown {Arabidopsi... 222 6e-58
BI272426 similar to GP|21554145|gb unknown {Arabidopsis thaliana... 154 1e-37
TC87045 similar to GP|21554145|gb|AAM63225.1 unknown {Arabidopsi... 111 1e-24
BF640903 weakly similar to GP|21464446|gb| RE73673p {Drosophila ... 96 8e-20
BE239842 similar to GP|17063179|gb| unknown protein {Arabidopsis... 57 3e-08
TC77752 weakly similar to GP|6728963|gb|AAF26961.1| putative N-a... 34 0.28
TC77953 similar to PIR|D84890|D84890 probable AT-hook DNA-bindin... 32 1.1
BQ138520 similar to SP|O59953|RL5_ 60S ribosomal protein L5 (CPR... 32 1.4
TC87487 similar to SP|P52780|SYQ_LUPLU Glutaminyl-tRNA synthetas... 32 1.4
AW585132 31 2.4
BI271537 similar to GP|19911209|db glucosyltransferase-13 {Vigna... 30 4.1
BQ137814 weakly similar to PIR|G02127|G021 fus-like protein - hu... 30 5.3
TC82510 weakly similar to GP|14532868|gb|AAK64116.1 unknown prot... 30 5.3
TC81393 similar to PIR|T00882|T00882 hypothetical protein At2g45... 30 5.3
>TC78607 similar to GP|20466746|gb|AAM20690.1 putative protein {Arabidopsis
thaliana}, partial (55%)
Length = 1734
Score = 516 bits (1330), Expect(3) = 0.0
Identities = 259/259 (100%), Positives = 259/259 (100%)
Frame = +3
Query: 404 HEGFLGALGAFMSYEKHGLDDLMVHQLVERFPMGAPYTGGKIHGPPLGDLNEKISWMEKF 463
HEGFLGALGAFMSYEKHGLDDLMVHQLVERFPMGAPYTGGKIHGPPLGDLNEKISWMEKF
Sbjct: 3 HEGFLGALGAFMSYEKHGLDDLMVHQLVERFPMGAPYTGGKIHGPPLGDLNEKISWMEKF 182
Query: 464 LQKGTEITAPVPMTPVAGTTGLGGFEVPLSKGSALRSDASALNVGVLHLVPTLEVFPLLA 523
LQKGTEITAPVPMTPVAGTTGLGGFEVPLSKGSALRSDASALNVGVLHLVPTLEVFPLLA
Sbjct: 183 LQKGTEITAPVPMTPVAGTTGLGGFEVPLSKGSALRSDASALNVGVLHLVPTLEVFPLLA 362
Query: 524 DPKLYEPNTIDLADPSELEYWLTILSEHLPDLVDKAVASEGGTDDAKRRGDAFARAFSAH 583
DPKLYEPNTIDLADPSELEYWLTILSEHLPDLVDKAVASEGGTDDAKRRGDAFARAFSAH
Sbjct: 363 DPKLYEPNTIDLADPSELEYWLTILSEHLPDLVDKAVASEGGTDDAKRRGDAFARAFSAH 542
Query: 584 LSRLMEEPSAYGKLGLANLLEMREECLREFQFVDAYRSIKQRENEASLAVLPDLFVELDS 643
LSRLMEEPSAYGKLGLANLLEMREECLREFQFVDAYRSIKQRENEASLAVLPDLFVELDS
Sbjct: 543 LSRLMEEPSAYGKLGLANLLEMREECLREFQFVDAYRSIKQRENEASLAVLPDLFVELDS 722
Query: 644 MDEETRLLTLIEGVLAANI 662
MDEETRLLTLIEGVLAANI
Sbjct: 723 MDEETRLLTLIEGVLAANI 779
Score = 418 bits (1075), Expect(3) = 0.0
Identities = 211/217 (97%), Positives = 214/217 (98%)
Frame = +2
Query: 676 KGTIIEIYRMSRNKMRRPWRVDDFDLFKERMLGTGDKKKAPHRRALLFVDNSGADIVLGM 735
K +++ YRMSRNKMRRPWRVDDFDLFKERMLGTGDKKKAPHRRALLFVDNSGADIVLGM
Sbjct: 821 KEQLLKFYRMSRNKMRRPWRVDDFDLFKERMLGTGDKKKAPHRRALLFVDNSGADIVLGM 1000
Query: 736 LPLARELLRRGTEVVLVANSLPALNDVTAMELPDIVAEAAKHCDILRRAAEAGGLLVDAM 795
LPLARELLRRGTEVVLVANSLPALNDVTAMELPDIVAEAAKHCDILRRAAEAGGLLVDAM
Sbjct: 1001 LPLARELLRRGTEVVLVANSLPALNDVTAMELPDIVAEAAKHCDILRRAAEAGGLLVDAM 1180
Query: 796 INTDSSKENSSSVPLMVVENGCGSPCIDLRQVSSELAAAAKDADLIILEGMGRSLHTNLY 855
INTDSSKENSSSVPLMVVENGCGSPCIDLRQVSSELAAAAKDADLIILEGMGRSLHTNLY
Sbjct: 1181 INTDSSKENSSSVPLMVVENGCGSPCIDLRQVSSELAAAAKDADLIILEGMGRSLHTNLY 1360
Query: 856 AQFKCDALKLAMVKNQRLAEKLIKGNIYDCICKYQPA 892
AQFKCDALKLAMVKNQRLAEKLIKGNIYDCICKYQPA
Sbjct: 1361 AQFKCDALKLAMVKNQRLAEKLIKGNIYDCICKYQPA 1471
Score = 45.8 bits (107), Expect(3) = 0.0
Identities = 19/19 (100%), Positives = 19/19 (100%)
Frame = +1
Query: 664 DWGSRACVDLYHKGTIIEI 682
DWGSRACVDLYHKGTIIEI
Sbjct: 784 DWGSRACVDLYHKGTIIEI 840
>TC78022 similar to GP|20466746|gb|AAM20690.1 putative protein {Arabidopsis
thaliana}, partial (31%)
Length = 1701
Score = 496 bits (1278), Expect = e-140
Identities = 250/411 (60%), Positives = 323/411 (77%), Gaps = 1/411 (0%)
Frame = +1
Query: 44 QLDVSKAEIQSNVEDKYPTILLPNQSDD-LSHLALDIGGSLIKLVYFSRHEGQSDDDKRK 102
++D + EIQ N + PN S + HLALDIGGSL KLVYF++ + D + +
Sbjct: 277 KMDQREFEIQPNSD--------PNPSTTHVYHLALDIGGSLNKLVYFTKDDDHFVDGEAE 432
Query: 103 KTLKERLGLSNGNRRSFPILGGRLHFVKFETGKINECLDFIHSKQLHRGLESRYSDTEAD 162
+ + S G+RR+ P+L G+L+F KFET KI++C+DFI + +LH G SR +
Sbjct: 433 ISPRNTSEKSKGSRRNHPVLKGKLNFKKFETSKIDDCIDFIKTMKLHPG-GSRQQENPGS 609
Query: 163 RNAIIKATGGGAYKFTDLFKEKLGVSLDKEDEMNCLVAGANFLLKAIRHEAFTHMEGQKE 222
+ IKATGGG+YK+ D FKE+LG++LDKEDEM+CLVAGANFLL+ + EA+T+M Q++
Sbjct: 610 QPISIKATGGGSYKYADPFKERLGINLDKEDEMDCLVAGANFLLEVVDREAYTYMGDQRQ 789
Query: 223 FVQIDQNDLFPYLLVNVGSGVSMIKVDGDGKFERVSGTNVGGGTYWGLGRLLTKCKSFDE 282
FVQIDQNDL+PYLLVN+GSGVSMIKV+G+GKFERVSGT++GGGT+WGLG+LLT+CKSFDE
Sbjct: 790 FVQIDQNDLYPYLLVNIGSGVSMIKVEGNGKFERVSGTSIGGGTFWGLGKLLTQCKSFDE 969
Query: 283 LLELSQKGDNRAIDMLVGDIYGGLDYSKIGLSASTIASSFGKTISEKKELQDYRPEDISL 342
LLELS +G+N+ +DMLVGDIYGG+DY+KIGLS++ IASSFGK +S+KKE +DYRPED++
Sbjct: 970 LLELSYQGNNKEVDMLVGDIYGGMDYAKIGLSSTAIASSFGKAMSDKKEREDYRPEDMAR 1149
Query: 343 SLLRMISYNIGQIAYLNALRFKLKRIFFGGFFIRGHAYTMDTLSFAVQYWSNGEAQAMFL 402
SLLRMIS NIGQI+YLNALRF LKRIFFGGFFI+ H +TMDTLS AV +WS GEA+AMFL
Sbjct: 1150SLLRMISNNIGQISYLNALRFGLKRIFFGGFFIQRHPFTMDTLSVAVNFWSKGEAKAMFL 1329
Query: 403 RHEGFLGALGAFMSYEKHGLDDLMVHQLVERFPMGAPYTGGKIHGPPLGDL 453
RHEGFLGA+GAFMS +KHGL +L+V Q ++ P + K++G G+L
Sbjct: 1330RHEGFLGAVGAFMSSDKHGLKELLVKQEAQQSPTKLSFAVDKMNGQLDGEL 1482
>BG646225 similar to GP|20466746|gb putative protein {Arabidopsis thaliana},
partial (20%)
Length = 623
Score = 394 bits (1013), Expect = e-110
Identities = 197/198 (99%), Positives = 197/198 (99%)
Frame = +3
Query: 44 QLDVSKAEIQSNVEDKYPTILLPNQSDDLSHLALDIGGSLIKLVYFSRHEGQSDDDKRKK 103
QLDVSKAEIQSNVEDKYPTILLPNQSDDLSHLALDIGGSLIKLVYFSRHEGQSDDDKRKK
Sbjct: 3 QLDVSKAEIQSNVEDKYPTILLPNQSDDLSHLALDIGGSLIKLVYFSRHEGQSDDDKRKK 182
Query: 104 TLKERLGLSNGNRRSFPILGGRLHFVKFETGKINECLDFIHSKQLHRGLESRYSDTEADR 163
TLKERLGLSNGNRRSFPILGGRLHFVKFETGKINECLDFIHSKQLHRGLESRYSDTEADR
Sbjct: 183 TLKERLGLSNGNRRSFPILGGRLHFVKFETGKINECLDFIHSKQLHRGLESRYSDTEADR 362
Query: 164 NAIIKATGGGAYKFTDLFKEKLGVSLDKEDEMNCLVAGANFLLKAIRHEAFTHMEGQKEF 223
NAIIKATGGGAYKFTDLFKEKLGVSLDKEDEMNCLVAGANFLLKAIRHEAFTHMEGQKEF
Sbjct: 363 NAIIKATGGGAYKFTDLFKEKLGVSLDKEDEMNCLVAGANFLLKAIRHEAFTHMEGQKEF 542
Query: 224 VQIDQNDLFPYLLVNVGS 241
VQIDQNDLFPYLLVNV S
Sbjct: 543 VQIDQNDLFPYLLVNVRS 596
>TC87044 similar to GP|21554145|gb|AAM63225.1 unknown {Arabidopsis
thaliana}, complete
Length = 1403
Score = 222 bits (565), Expect = 6e-58
Identities = 131/356 (36%), Positives = 204/356 (56%), Gaps = 2/356 (0%)
Frame = +3
Query: 536 ADPSELEYWLTILSEHLPDLVDKAVASEGGTDDAKRRGDAFARAFSAHLSRLMEEPSAYG 595
A P+E+ W+ + +P +A S+ +A + + FA+ +S L ++P ++G
Sbjct: 195 ATPTEIS-WINLFLNSIPSFKKRA-ESDTSVPNAAEKAEKFAQRYSDILEDFKKDPESHG 368
Query: 596 -KLGLANLLEMREECLREFQFVDAYRSIKQRENEASLAVLPDLFVELDSMDEET-RLLTL 653
L +RE LRE F D ++ +K EN ++++ ++ D++++E R+ L
Sbjct: 369 GPPDCIVLCRLREIILRELGFQDIFKKVKDEENAKAISLFENVVRVNDAIEDEVKRVENL 548
Query: 654 IEGVLAANIFDWGSRACVDLYHKGTIIEIYRMSRNKMRRPWRVDDFDLFKERMLGTGDKK 713
++G+ A NIFD GS +++ K + +N + RPW +DD + FK + + KK
Sbjct: 549 VKGIFAGNIFDLGSAQLAEVFSKDGM-SFLASCQNLVTRPWVIDDLEAFKVKWIKNSWKK 725
Query: 714 KAPHRRALLFVDNSGADIVLGMLPLARELLRRGTEVVLVANSLPALNDVTAMELPDIVAE 773
++FVDNSGADI+LG+LP ARELLRRG++VVL AN LP++NDVT EL +I+++
Sbjct: 726 ------VIIFVDNSGADIILGILPFARELLRRGSQVVLAANDLPSINDVTYSELVEIISK 887
Query: 774 AAKHCDILRRAAEAGGLLVDAMINTDSSKENSSSVPLMVVENGCGSPCIDLRQVSSELAA 833
E G L+ S+ L++ +G P IDL +VS ELA
Sbjct: 888 LKD---------EEGNLV------------GVSTSNLLIANSGNDLPVIDLARVSQELAY 1004
Query: 834 AAKDADLIILEGMGRSLHTNLYAQFKCDALKLAMVKNQRLAEKLIKGNIYDCICKY 889
A DADL++LEGMGR + TNLYAQFKCD+LK+ MVK+ +A+ + G +YDC+ KY
Sbjct: 1005LASDADLVVLEGMGRGIETNLYAQFKCDSLKIGMVKHPEVAQ-FLGGRLYDCVLKY 1169
>BI272426 similar to GP|21554145|gb unknown {Arabidopsis thaliana}, partial
(58%)
Length = 646
Score = 154 bits (390), Expect = 1e-37
Identities = 92/242 (38%), Positives = 136/242 (56%), Gaps = 1/242 (0%)
Frame = +3
Query: 617 DAYRSIKQRENEASLAVLPDLFVELDSMDEE-TRLLTLIEGVLAANIFDWGSRACVDLYH 675
D ++ +K EN ++++ ++ D++++E RL L+ G+ A N+FD GS + +
Sbjct: 3 DIFKKVKAEENAKAISLFENVVRLNDAIEDEGNRLENLVRGIFAGNVFDLGSPQLAEAFS 182
Query: 676 KGTIIEIYRMSRNKMRRPWRVDDFDLFKERMLGTGDKKKAPHRRALLFVDNSGADIVLGM 735
+ + N + RPW +DD D K R K ++ ++FVDNSGADI+LG+
Sbjct: 183 RDGM-SFSATCENLLPRPWIIDDLDTLKIRW------SKKSWKKVIIFVDNSGADIILGI 341
Query: 736 LPLARELLRRGTEVVLVANSLPALNDVTAMELPDIVAEAAKHCDILRRAAEAGGLLVDAM 795
LP ARELLRRG++VVL AN LP++NDVT EL +I+++ E G LL
Sbjct: 342 LPFARELLRRGSQVVLAANDLPSINDVTYSELIEIISKLKD---------EEGRLL---- 482
Query: 796 INTDSSKENSSSVPLMVVENGCGSPCIDLRQVSSELAAAAKDADLIILEGMGRSLHTNLY 855
S+ L++ +G P IDL +VS ELA+ D DL+ILEGMG + TNLY
Sbjct: 483 --------GVSTSKLLIANSGNDLPVIDLTKVSQELASLTSDVDLVILEGMGXGIKTNLY 638
Query: 856 AQ 857
AQ
Sbjct: 639 AQ 644
>TC87045 similar to GP|21554145|gb|AAM63225.1 unknown {Arabidopsis
thaliana}, partial (39%)
Length = 717
Score = 111 bits (277), Expect = 1e-24
Identities = 77/221 (34%), Positives = 106/221 (47%), Gaps = 58/221 (26%)
Frame = +3
Query: 727 SGADIVLGMLPLARELLRRGTEVVLVANSLPALNDVTAMELPDIVAEAAKHCDILRRAAE 786
SGADI+LG+LP ARELLRRG++VVL AN LP++NDVT EL +I+++ E
Sbjct: 6 SGADIILGILPFARELLRRGSQVVLAANDLPSINDVTYSELVEIISKLKD---------E 158
Query: 787 AGGLLVDAMINTDSSKENSSSVPLMVVENGCGSPCIDLRQVSSELAAAAKDADLI----- 841
G L+ S+ L++ +G P IDL +VS ELA A DADL+
Sbjct: 159 EGNLV------------GVSTSNLLIANSGNDLPVIDLARVSQELAYLASDADLVVLEGM 302
Query: 842 ILEGMG-----------------------------------------------------R 848
+L+G+G R
Sbjct: 303 VLDGLGP*PVVAPGAF*TYRHVVSEPQFDLMGDH*WILAVLSNVGDSRLG*LLEFRRVGR 482
Query: 849 SLHTNLYAQFKCDALKLAMVKNQRLAEKLIKGNIYDCICKY 889
+ TNLYAQFKCD+LK+ MVK+ +A+ + G +YDC+ KY
Sbjct: 483 GIETNLYAQFKCDSLKIGMVKHPEVAQ-FLGGRLYDCVLKY 602
>BF640903 weakly similar to GP|21464446|gb| RE73673p {Drosophila
melanogaster}, partial (19%)
Length = 686
Score = 95.5 bits (236), Expect = 8e-20
Identities = 58/179 (32%), Positives = 91/179 (50%), Gaps = 2/179 (1%)
Frame = +3
Query: 521 LLADPKLYEPNTIDL-ADPSELEYWLTILSEHLPDLVDKAVASEGGTDDAKRRGDAFARA 579
+L +P Y P+T+DL D EYWL + V+ A+ + + R F
Sbjct: 147 ILLEPDKYNPDTLDLLVDEEAREYWLNTCEKLTEKYVNFALLNNEDPT-VEIRALKFKTC 323
Query: 580 FSAHLSRLMEEPSAYGKLGLANLLEMREECLREFQFVDAYRSIKQRENEASLAVLPDLFV 639
+ L L P A+G+L + LL++ E CLR F D ++ K+ EN+++LA L
Sbjct: 324 YVEALKELRINPLAHGQLTIRLLLDINETCLRSQGFFDLWKKQKKYENDSALATLATRIA 503
Query: 640 ELDSM-DEETRLLTLIEGVLAANIFDWGSRACVDLYHKGTIIEIYRMSRNKMRRPWRVD 697
ELD++ D+ R + +I GVLA N+FDWG++A + + G +Y +RPW D
Sbjct: 504 ELDALPDDRQRWIEIIRGVLAGNMFDWGAQAVTSILNSG----LYEALEKIXKRPWXYD 668
>BE239842 similar to GP|17063179|gb| unknown protein {Arabidopsis thaliana},
partial (12%)
Length = 400
Score = 57.0 bits (136), Expect = 3e-08
Identities = 24/43 (55%), Positives = 34/43 (78%)
Frame = +2
Query: 847 GRSLHTNLYAQFKCDALKLAMVKNQRLAEKLIKGNIYDCICKY 889
GR + TNLYAQFKCD++K+AMVK+ +AE ++ +YDC+ KY
Sbjct: 2 GRGIETNLYAQFKCDSMKIAMVKHPEVAE-FLESRMYDCVIKY 127
>TC77752 weakly similar to GP|6728963|gb|AAF26961.1| putative
N-acetlytransferase {Arabidopsis thaliana}, partial
(82%)
Length = 1178
Score = 33.9 bits (76), Expect = 0.28
Identities = 27/77 (35%), Positives = 36/77 (46%)
Frame = +3
Query: 345 LRMISYNIGQIAYLNALRFKLKRIFFGGFFIRGHAYTMDTLSFAVQYWSNGEAQAMFLRH 404
L +IS+NI I + FK R G +FI G Y D+ F V Y + G + L+
Sbjct: 630 LHVISFNISAINLYKKMSFKCVRKLQGFYFINGQHY--DSFLF-VHYVNGGRSPCSPLQ- 797
Query: 405 EGFLGALGAFMSYEKHG 421
L AF+SY K G
Sbjct: 798 -----LLSAFVSYMKSG 833
>TC77953 similar to PIR|D84890|D84890 probable AT-hook DNA-binding protein
[imported] - Arabidopsis thaliana, partial (57%)
Length = 1695
Score = 32.0 bits (71), Expect = 1.1
Identities = 16/54 (29%), Positives = 32/54 (58%)
Frame = +1
Query: 9 QQQQQVLGIDESEGHNNTNNMAPTGNPIHRSSSRPQLDVSKAEIQSNVEDKYPT 62
QQ+ + I+ + +NN N + T ++ SS++PQ+ ++KAE+ + V+ T
Sbjct: 457 QQEIFIFTINTNSSNNNNNTINST---LYSSSNKPQIKMNKAEVAAAVDSTSQT 609
>BQ138520 similar to SP|O59953|RL5_ 60S ribosomal protein L5 (CPR4).
{Neurospora crassa}, partial (63%)
Length = 584
Score = 31.6 bits (70), Expect = 1.4
Identities = 18/69 (26%), Positives = 37/69 (53%), Gaps = 6/69 (8%)
Frame = -3
Query: 239 VGSGVSMIKVD------GDGKFERVSGTNVGGGTYWGLGRLLTKCKSFDELLELSQKGDN 292
+ +G S+++++ G+ K + G VG +G L +S ++L+ + +GDN
Sbjct: 345 LNTGESLVELELLEDTAGEQKTSGIGGRPVGQTVGDAIGLELVSVRSHEDLVATNLRGDN 166
Query: 293 RAIDMLVGD 301
+ D+LVG+
Sbjct: 165 LSDDVLVGE 139
>TC87487 similar to SP|P52780|SYQ_LUPLU Glutaminyl-tRNA synthetase (EC
6.1.1.18) (Glutamine--tRNA ligase) (GLNRS). [Yellow
lupine], partial (97%)
Length = 2844
Score = 31.6 bits (70), Expect = 1.4
Identities = 23/71 (32%), Positives = 37/71 (51%), Gaps = 2/71 (2%)
Frame = +2
Query: 508 GVLHLV--PTLEVFPLLADPKLYEPNTIDLADPSELEYWLTILSEHLPDLVDKAVASEGG 565
GVLH V P EV PL + +L++ + +P+EL+ WL L+ H +++ A
Sbjct: 2165 GVLHWVAQPAHEVDPLKVEVRLFDRLFLS-ENPAELDNWLGDLNPHSKEVIPNAFGLPSL 2341
Query: 566 TDDAKRRGDAF 576
D + GD+F
Sbjct: 2342 RD--AKVGDSF 2368
>AW585132
Length = 541
Score = 30.8 bits (68), Expect = 2.4
Identities = 42/155 (27%), Positives = 57/155 (36%), Gaps = 5/155 (3%)
Frame = +1
Query: 303 YGGLDYSKIGLSASTIASSFGKTISEKKELQDYRPEDISLSLLRMISYNIGQIAYLNALR 362
+GG D+ K G S GK +KKE +D+R ED R N G + N
Sbjct: 55 HGGTDHVKTGSLLIVTVLSPGKVTLDKKE*EDHRFEDTV*LSSRTTLNNTG*TLHPNVSC 234
Query: 363 FKLKRIFFGGFFIRGHAYTMDTLSFAVQYWSNGEAQAMFLRHEGFLGALGAFMSYEKHGL 422
KR GG + D GE + +EG G G S +K+G
Sbjct: 235 PGSKRTPPGGPPT*RYFPITDISLAVANPRLKGEVKVTPPWYEGPSGVAGVPTSLDKYGS 414
Query: 423 DDLMVH-----QLVERFPMGAPYTGGKIHGPPLGD 452
+ L++ P+ A T G G P GD
Sbjct: 415 KEPPAK*EV**SLIKPLPV-ADKTNG*SDGEPTGD 516
>BI271537 similar to GP|19911209|db glucosyltransferase-13 {Vigna angularis},
partial (34%)
Length = 652
Score = 30.0 bits (66), Expect = 4.1
Identities = 22/69 (31%), Positives = 33/69 (46%)
Frame = +2
Query: 459 WMEKFLQKGTEITAPVPMTPVAGTTGLGGFEVPLSKGSALRSDASALNVGVLHLVPTLEV 518
++E+ +KG IT+ VP + +GGF S L S V+H VP L
Sbjct: 272 FLERTKEKGFVITSWVPQIQILSHNSVGGFLTHCGWNSTLES--------VVHGVP-LIT 424
Query: 519 FPLLADPKL 527
+PL A+ K+
Sbjct: 425 WPLFAEQKM 451
>BQ137814 weakly similar to PIR|G02127|G021 fus-like protein - human
(fragment), partial (8%)
Length = 1050
Score = 29.6 bits (65), Expect = 5.3
Identities = 15/37 (40%), Positives = 24/37 (64%)
Frame = +3
Query: 240 GSGVSMIKVDGDGKFERVSGTNVGGGTYWGLGRLLTK 276
G+G+ + G+G+ +G +GGGT+ G GRLLT+
Sbjct: 156 GTGLFAERGAGEGRAPAPAG-EIGGGTWRGAGRLLTE 263
>TC82510 weakly similar to GP|14532868|gb|AAK64116.1 unknown protein
{Arabidopsis thaliana}, partial (31%)
Length = 1716
Score = 29.6 bits (65), Expect = 5.3
Identities = 26/87 (29%), Positives = 40/87 (45%), Gaps = 3/87 (3%)
Frame = +1
Query: 422 LDDLMVHQLVERFPMGAPYTGGKIHGPPLGDLNEKISWMEKFLQKGTEITAPVPMTPVAG 481
LD + + L+ P G Y L D+N + +++ FL++GT AP+ M VA
Sbjct: 889 LDQVTLDNLLVPSPYGINY---------LYDVNLVLRFLKAFLRQGTGAVAPIRMRKVAS 1041
Query: 482 TTGLGGFEV---PLSKGSALRSDASAL 505
L E+ P K S + A+AL
Sbjct: 1042LIDLYIAEIAPDPCLKTSKFLALATAL 1122
>TC81393 similar to PIR|T00882|T00882 hypothetical protein At2g45690
[imported] - Arabidopsis thaliana, partial (35%)
Length = 716
Score = 29.6 bits (65), Expect = 5.3
Identities = 15/34 (44%), Positives = 21/34 (61%)
Frame = -2
Query: 229 NDLFPYLLVNVGSGVSMIKVDGDGKFERVSGTNV 262
N+L P+LLV+ S + IK D FE+ SGT +
Sbjct: 715 NNLSPHLLVSCFSNHNSIKFDMSSSFEKNSGTKL 614
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.319 0.137 0.397
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 23,013,787
Number of Sequences: 36976
Number of extensions: 285097
Number of successful extensions: 1293
Number of sequences better than 10.0: 34
Number of HSP's better than 10.0 without gapping: 1275
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1285
length of query: 892
length of database: 9,014,727
effective HSP length: 105
effective length of query: 787
effective length of database: 5,132,247
effective search space: 4039078389
effective search space used: 4039078389
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 63 (28.9 bits)
Medicago: description of AC131239.17


