

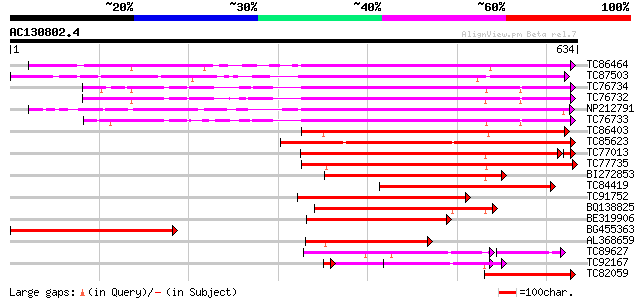
BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC130802.4 + phase: 0
(634 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC86464 similar to GP|2098711|gb|AAB57670.1| pectinesterase {Cit... 345 4e-95
TC87503 similar to SP|Q43111|PME3_PHAVU Pectinesterase 3 precurs... 335 2e-92
TC76734 similar to PIR|T06468|T06468 pectinesterase (EC 3.1.1.11... 330 1e-90
TC76732 homologue to PIR|T06468|T06468 pectinesterase (EC 3.1.1.... 322 3e-88
NP212791 NP212791|AJ249611.1|CAB65291.1 pectin methyl-esterase PEF1 322 3e-88
TC76733 similar to PIR|T06468|T06468 pectinesterase (EC 3.1.1.11... 315 4e-86
TC86403 similar to PIR|B86158|B86158 hypothetical protein AAF028... 306 2e-83
TC85623 similar to GP|13605696|gb|AAK32841.1 At2g45220/F4L23.27 ... 300 1e-81
TC77013 similar to GP|7025485|gb|AAF35897.1| pectin methylestera... 295 4e-80
TC77735 similar to GP|14334646|gb|AAK59501.1 putative pectineste... 293 1e-79
BI272853 weakly similar to PIR|T04359|T04 pectin methylesterase-... 213 2e-55
TC84419 similar to SP|Q42920|PME_MEDSA Pectinesterase precursor ... 209 2e-54
TC91752 similar to GP|6630559|gb|AAF19578.1| putative pectineste... 203 1e-52
BQ138825 similar to PIR|T49922|T49 pectin methylesterase-like pr... 198 6e-51
BE319906 similar to SP|Q43062|PME_ Pectinesterase PPE8B precurso... 185 4e-47
BG455363 weakly similar to PIR|T52331|T523 pectinesterase (EC 3.... 155 3e-38
AL368659 similar to PIR|B86158|B86 hypothetical protein AAF02886... 149 3e-36
TC89627 weakly similar to GP|17529290|gb|AAL38872.1 putative pec... 119 2e-33
TC92167 similar to PIR|T05203|T05203 pectinesterase homolog F4I1... 115 2e-28
TC82059 similar to GP|19424045|gb|AAL87311.1 unknown protein {Ar... 113 2e-25
>TC86464 similar to GP|2098711|gb|AAB57670.1| pectinesterase {Citrus
sinensis}, partial (85%)
Length = 1993
Score = 345 bits (884), Expect = 4e-95
Identities = 222/626 (35%), Positives = 316/626 (50%), Gaps = 15/626 (2%)
Frame = +1
Query: 22 KKKILIGVTSVVLIACVIAAVTFVIVKRSGPDHNNNDKKPVQNAPPEPERVDKYSRLVTM 81
KKK+LI + + +LI I A+ K S NN+ + + K +
Sbjct: 97 KKKLLISLFTTLLIVASIVAIVATTTKNSNKSKNNSIASSSLSLSHHSHAILKSA----- 261
Query: 82 LCSHSEYKEKCVTTLKEALKKDPKLKEPKGLLMVFMLVAKNEINNAFNKTANL---KFAS 138
C+ + Y E C + + K+ K ++ + + + + + + L K +
Sbjct: 262 -CTTTLYPELCFSAISSEPNITHKITNHKDVISLSLNITTRAVEHNYFTVEKLLLRKSLT 438
Query: 139 KEEKGAYEDCKQLFEDAKEEMGFSITEVGQLDISKLASKEAE-LNNWLSAVISYQDTCSD 197
K EK A DC + ++ +E+ + ++ K + A+ L +S+ I+ Q TC D
Sbjct: 439 KREKIALHDCLETIDETLDELKEAQNDLVLYPSKKTLYQHADDLKTLISSAITNQVTCLD 618
Query: 198 GFPEGELKKKMEMIFAESR----QLLSNSLAVVSQVSQIVNAFQGGLSGFKLPWGKSDAH 253
GF + K++ + E + + SN+LA+ N ++ F+
Sbjct: 619 GFSHDDADKEVRKVLQEGQIHVEHMCSNALAMTK------NMTDKDIAEFE--------- 753
Query: 254 APAPDADTDAVADDDED--LADAPDGAPDADQPIFEAPIGAPGAAPIGAPRVDAPPSWAA 311
T+ V +++ L + +G +G P G R+
Sbjct: 754 ------QTNMVLGSNKNRKLLEEENG------------VGWPEWISAGDRRL-------- 855
Query: 312 PAVDLPGSTEKPTPNVTVAKDGSGDFKTISEALAAIPQTYKGRYVVYVKEGVYDETVTVT 371
L GST K +V VA DGSG+FKT+SEA+AA P RYV+ +K GVY E V V
Sbjct: 856 ----LQGSTVKA--DVVVAADGSGNFKTVSEAVAAAPLKSSKRYVIKIKAGVYKENVEVP 1017
Query: 372 KKMVNLTMYGDGGLKSIITGNKNFVDGVRTFQTASFVVLGDGFVGRDMGFRNTAGAIKEQ 431
KK N+ GDG +IITG++N VDG TF +A+ ++G F+ RD+ F+NTAG K Q
Sbjct: 1018KKKTNIMFLGDGRTNTIITGSRNVVDGSTTFHSATVAIVGGNFLARDITFQNTAGPAKHQ 1197
Query: 432 AVAARVQADCSIFVNCNFEGYQDTLYAQTHRQFYRDCVISGTIDFIFGHASAVFQNCQLV 491
AVA RV AD S F NC+ YQDTLY +RQF+ +C ISGT+DFIFG+++ VFQNC +
Sbjct: 1198AVALRVGADLSAFYNCDIIAYQDTLYVHNNRQFFVNCFISGTVDFIFGNSAVVFQNCDIH 1377
Query: 492 LRKPLDNQKNIITANGRIDSKSNTAFVLQKCVIKGEDDLPSTTKN---YIGRPWKEYSRT 548
R+P QKN++TA GR+D NT V+QKC I DL N Y+GRPWKEYSRT
Sbjct: 1378ARRPNSGQKNMVTAQGRVDPNQNTGIVIQKCRIGATKDLEGVKGNFPTYLGRPWKEYSRT 1557
Query: 549 IIMESDIPALIQPEGWLPWEGDFALKTLYYGEYDNVGAGAKTDARVKWIGRKDI-KRGEA 607
+ M+S I +I P GW W G+FAL TL Y EY N G GA T RV W G K I EA
Sbjct: 1558VFMQSSISDVIDPVGWHEWNGNFALNTLVYREYQNTGPGAGTSKRVTWKGFKVITSAAEA 1737
Query: 608 LTYTVEPFLDG-SWINGTGVPAHLGL 632
+ T F+ G SW+ TG P LGL
Sbjct: 1738QSSTPGNFIGGSSWLGSTGFPFSLGL 1815
>TC87503 similar to SP|Q43111|PME3_PHAVU Pectinesterase 3 precursor (EC
3.1.1.11) (Pectin methylesterase 3) (PE 3)., partial
(93%)
Length = 2088
Score = 335 bits (860), Expect = 2e-92
Identities = 212/640 (33%), Positives = 340/640 (53%), Gaps = 15/640 (2%)
Frame = +2
Query: 2 AFEDFDLVSE-RRKANAKQHLKKKILIGVTSVVLIACVIAAVTFVIVKRSGPDHNNNDKK 60
+F+ + V E ++A K+ K+ +I ++S++L+A +IAAV +++ + +N +
Sbjct: 74 SFKGYGKVDEIEQQAFQKKTRKRITIIIISSIILVAVIIAAVAGILIHK-----HNTESS 238
Query: 61 PVQNAPPEPERVDKYSRLVTMLCSHSEYKEKCVTTLKEALKKDPKLKEPKGLLMVFMLVA 120
N+ P E S + +C ++Y C +++ D +P+ L + + VA
Sbjct: 239 SSPNSLPNTELTPATS--LKAVCESTQYPNSCFSSISSL--PDSNTTDPEQLFKLSLKVA 406
Query: 121 KNEINNAFNKTANLKFASKEEKGAYEDCKQLFEDAKEEMGFSITEVGQLDISKLAS--KE 178
+E++ + K K A C + D+ + + S++ + +D K+ S K
Sbjct: 407 IDELSKLSLTRFSEKATEPRVKKAIGVCDNVLADSLDRLNDSMSTI--VDGGKMLSPAKI 580
Query: 179 AELNNWLSAVISYQDTCSDGFPEGE------LKKKMEMIFAESRQLLSNSLAVVSQVSQI 232
++ WLSA ++ DTC D E + ++E I S + SNSLA+VS+V ++
Sbjct: 581 RDVETWLSAALTDHDTCLDAVGEVNSTAARGVIPEIERIMRNSTEFASNSLAIVSKVIRL 760
Query: 233 VNAFQGGLSGFKLPWGKSDAHAPAPDADTDAVADDDEDLADAPDGAPDADQPIFEAPIGA 292
LS F++ S+ H L + P+ A++ + +
Sbjct: 761 -------LSNFEV----SNHHRRL--------------LGEFPEWLGTAERRLLATVVN- 862
Query: 293 PGAAPIGAPRVDAPPSWAAPAVDLPGSTEKPTPNVTVAKDGSGDFKTISEALAAIPQTYK 352
+ P+ VAKDGSG +KTI EAL + +
Sbjct: 863 -----------------------------ETVPDAVVAKDGSGQYKTIGEALKLVKKKSL 955
Query: 353 GRYVVYVKEGVYDETVTVTKKMVNLTMYGDGGLKSIITGNKNFVDGVRTFQTASFVVLGD 412
R+VVYVK+GVY E + + K N+ +YGDG +++++G++N++DG TF+TA+F V G
Sbjct: 956 QRFVVYVKKGVYVENIDLDKNTWNVMIYGDGMTETVVSGSRNYIDGTPTFETATFAVKGK 1135
Query: 413 GFVGRDMGFRNTAGAIKEQAVAARVQADCSIFVNCNFEGYQDTLYAQTHRQFYRDCVISG 472
GF+ +D+ F NTAGA K QAVA R +D S+F C+F GYQDTLYA ++RQFYRDC I+G
Sbjct: 1136GFIAKDIQFLNTAGASKHQAVAMRSGSDQSVFYRCSFVGYQDTLYAHSNRQFYRDCDITG 1315
Query: 473 TIDFIFGHASAVFQNCQLVLRKPLDNQKNIITANGRIDSKSNTAFVLQK---CVIKGEDD 529
TIDFIFG+A+AVFQNC+++ R+P+ NQ N ITA G+ D N+ V+QK + G++
Sbjct: 1316TIDFIFGNAAAVFQNCKIMPRQPMSNQFNTITAQGKKDPNQNSGIVIQKSTFTTLPGDNL 1495
Query: 530 LPSTTKNYIGRPWKEYSRTIIMESDIPALIQPEGWLPWEGDF-ALKTLYYGEYDNVGAGA 588
+ T Y+GRPWK++S TIIM+S+I + ++P GW+ W + ++ Y EY N G GA
Sbjct: 1496IAPT---YLGRPWKDFSTTIIMKSEIGSFLKPVGWISWVANVEPPSSILYAEYQNTGPGA 1666
Query: 589 KTDARVKWIGRKDIKRGE-ALTYTVEPFLDG-SWINGTGV 626
RVKW G K E A+ +TV+ F+ G W+ V
Sbjct: 1667DVPGRVKWAGYKPALGDEDAIKFTVDSFIQGPEWLPSASV 1786
>TC76734 similar to PIR|T06468|T06468 pectinesterase (EC 3.1.1.11) precursor
- garden pea, complete
Length = 1981
Score = 330 bits (845), Expect = 1e-90
Identities = 210/568 (36%), Positives = 294/568 (50%), Gaps = 17/568 (2%)
Frame = +3
Query: 82 LCSHSEYKEKCVTTLKEALK-------KDPKLKEPKGLLMVFMLVAKNEINNAFNKTANL 134
+C H+ C+T + E ++ KD KL L+ + + I A + TAN+
Sbjct: 201 VCEHAVDTNSCLTHVAEVVQGSTLDNTKDHKLST----LISLLTKSTTHIRKAMD-TANV 365
Query: 135 ---KFASKEEKGAYEDCKQLFEDAKEEMGFSITEVGQLDISKLASKEAELNNWLSAVISY 191
+ S E+ A C++L + E + S+ + + ++ + + + WLS+V++
Sbjct: 366 IKRRINSPREENALNVCEKLMNLSMERVWDSVLTLTKDNMDS----QQDAHTWLSSVLTN 533
Query: 192 QDTCSDGFPEGELKKKMEMIFAESRQLLSNSLAVVSQVSQIVNAFQGGLSGFKLPWGKSD 251
TC DG EG + ME + +SLAV+ V
Sbjct: 534 HATCLDGL-EGTSRAVMENDIQDLIARARSSLAVLVAV---------------------- 644
Query: 252 AHAPAPDADTDAVADDDEDLADAPDGAPDADQPIFEAPIGAPGAAPIGAPRVDAPPSWAA 311
P D D D+ + D P D+ + E+ +G A
Sbjct: 645 ----LPPKDHDEFIDESLN-GDFPSWVTSKDRRLLESSVGDVKA---------------- 761
Query: 312 PAVDLPGSTEKPTPNVTVAKDGSGDFKTISEALAAIPQTYKGRYVVYVKEGVYDETVTVT 371
NV VAKDGSG FKT++EA+A+ P RYV+YVK+G+Y E V +
Sbjct: 762 --------------NVVVAKDGSGKFKTVAEAVASAPNKGTARYVIYVKKGIYKENVEIA 899
Query: 372 KKMVNLTMYGDGGLKSIITGNKNFVDGVRTFQTASFVVLGDGFVGRDMGFRNTAGAIKEQ 431
N+ + GDG +IITG+ N+VDG TFQTA+ +GD F+ +D+GF+NTAG K Q
Sbjct: 900 SSKTNVMLLGDGMDATIITGSLNYVDGTGTFQTATVAAVGDWFIAQDIGFQNTAGPQKHQ 1079
Query: 432 AVAARVQADCSIFVNCNFEGYQDTLYAQTHRQFYRDCVISGTIDFIFGHASAVFQNCQLV 491
AVA RV +D S+ C + +QDTLYA T+RQFYRD I+GTIDFIFG A+ V Q C+LV
Sbjct: 1080AVALRVGSDRSVINRCKIDAFQDTLYAHTNRQFYRDSFITGTIDFIFGDAAVVLQKCKLV 1259
Query: 492 LRKPLDNQKNIITANGRIDSKSNTAFVLQKCVIKGEDDLP---STTKNYIGRPWKEYSRT 548
RKP+ NQ N++TA GRID NTA +Q+C + DL + K Y+GRPWK+YSRT
Sbjct: 1260ARKPMANQNNMVTAQGRIDPNQNTATSIQQCDVIPSTDLKPVIGSVKTYLGRPWKKYSRT 1439
Query: 549 IIMESDIPALIQPEGWLPWEG---DFALKTLYYGEYDNVGAGAKTDARVKWIGRKDIKRG 605
++M+S + A I P GW W+ DF L+TLYYGEY N G GA T RVKW G I
Sbjct: 1440VVMQSLLGAHIDPTGWAEWDAASKDF-LQTLYYGEYMNSGPGAGTSKRVKWPGYHIINTA 1616
Query: 606 EALTYTVEPFLDGS-WINGTGVPAHLGL 632
EA +TV + G+ W+ TGV GL
Sbjct: 1617EANKFTVAQLIQGNVWLKNTGVAFIAGL 1700
>TC76732 homologue to PIR|T06468|T06468 pectinesterase (EC 3.1.1.11)
precursor - garden pea, complete
Length = 1860
Score = 322 bits (825), Expect = 3e-88
Identities = 205/564 (36%), Positives = 298/564 (52%), Gaps = 13/564 (2%)
Frame = +2
Query: 82 LCSHSEYKEKCVTTLKEALKKDP--KLKEPKGLLMVFMLVAKNEINNAFNKTANL---KF 136
LC H+ + C+T + E ++ K+ K +V +L TAN+ +
Sbjct: 194 LCEHALDTKSCLTHVSEVVQGSTLSNTKDHKLSTLVSLLTKSTAHIRKAMDTANVIKRRV 373
Query: 137 ASKEEKGAYEDCKQLFEDAKEEMGFSITEVGQLDISKLASKEAELNNWLSAVISYQDTCS 196
S E+ A DC++L + + + + S+ + + +I + + + WLS+V++ TC
Sbjct: 374 NSPREEIALNDCEELMDLSMDRVWDSVLTLTKNNIDS----QHDAHTWLSSVLTNHATCL 541
Query: 197 DGFPEGELKKKMEMIFAESRQLLSNSLAVVSQVSQIVNAFQGGLSGFKLPWGKSDAHAPA 256
DG EG + ME + +SLAV+ V LP
Sbjct: 542 DGL-EGSSRVVMESDLHDLISRARSSLAVLVSV---------------LP---------- 643
Query: 257 PDADTDAVADDDEDLADAPDGAPDADQPIFEAPIGAPGAAPIGAPRVDAPPSWAAPAVDL 316
P A+ + D++ D P D+ + E+ +G A
Sbjct: 644 PKANDGFI--DEKLNGDFPSWVTSKDRRLLESSVGDIKA--------------------- 754
Query: 317 PGSTEKPTPNVTVAKDGSGDFKTISEALAAIPQTYKGRYVVYVKEGVYDETVTVTKKMVN 376
NV VA+DGSG FKT+++A+A+ P K RYV+YVK+G Y E + + KK N
Sbjct: 755 ---------NVVVAQDGSGKFKTVAQAVASAPDNGKTRYVIYVKKGTYKENIEIGKKKTN 907
Query: 377 LTMYGDGGLKSIITGNKNFVDGVRTFQTASFVVLGDGFVGRDMGFRNTAGAIKEQAVAAR 436
+ + GDG +IITG+ NF+DG TF++A+ +GDGF+ +D+ F+NTAG K QAVA R
Sbjct: 908 VMLVGDGMDATIITGSLNFIDGTTTFKSATVAAVGDGFIAQDIRFQNTAGPQKHQAVALR 1087
Query: 437 VQADCSIFVNCNFEGYQDTLYAQTHRQFYRDCVISGTIDFIFGHASAVFQNCQLVLRKPL 496
V AD S+ C + +QDTLYA ++RQFYRD I+GT+DFIFG+A+ VFQ +L RKP+
Sbjct: 1088VGADQSVINRCKIDAFQDTLYAHSNRQFYRDSYITGTVDFIFGNAAVVFQKSKLAARKPM 1267
Query: 497 DNQKNIITANGRIDSKSNTAFVLQKCVIKGEDDL---PSTTKNYIGRPWKEYSRTIIMES 553
NQKN++TA GR D NTA +Q+C + DL + K Y+GRPWK+YSRT++++S
Sbjct: 1268ANQKNMVTAQGREDPNQNTATSIQQCDVIPSSDLKPVQGSIKTYLGRPWKKYSRTVVLQS 1447
Query: 554 DIPALIQPEGWLPWEG---DFALKTLYYGEYDNVGAGAKTDARVKWIGRKDIKR-GEALT 609
+ I P GW W+ DF L+TLYYGEY N GAGA T RV W G IK EA
Sbjct: 1448VVDGHIDPAGWAEWDAASKDF-LQTLYYGEYMNSGAGAGTGKRVTWPGYHIIKNAAEASK 1624
Query: 610 YTVEPFLDGS-WINGTGVPAHLGL 632
+TV + G+ W+ TGV GL
Sbjct: 1625FTVTQLIQGNVWLKNTGVAFIEGL 1696
>NP212791 NP212791|AJ249611.1|CAB65291.1 pectin methyl-esterase PEF1
Length = 1698
Score = 322 bits (825), Expect = 3e-88
Identities = 216/623 (34%), Positives = 308/623 (48%), Gaps = 13/623 (2%)
Frame = +1
Query: 22 KKKILIGVTSVVLIACVIAAVTFVIVKRSGPDHNNNDKKPVQNAPPEPERVDKYSRLVTM 81
KK L+GV+ ++L+A V V V + + G + K + N+ + V M
Sbjct: 40 KKHALLGVSCILLVAMV--GVVAVSLTKGG---DGEQKAHISNS----------QKNVDM 174
Query: 82 LCSHSEYKEKCVTTLKEALKKDPKLKEPKGLLMVFMLVAKNEINNA--FNKTANLKFASK 139
LC +++KE C TL++A + K KG L + INN+ + + A
Sbjct: 175 LCQSTKFKETCHKTLEKASFSNMK-NRIKGALGATEEELRKHINNSALYQELAT----DS 339
Query: 140 EEKGAYEDCKQLFEDAKEEMGFSITEVGQLDISKLASKEAELNNWLSAVISYQDTCSDGF 199
K A E C ++ + A + + S+ + Q D KL+ ++ WL+ +S+Q TC DGF
Sbjct: 340 MTKQAMEICNEVLDYAVDGIHKSVGTLDQFDFHKLSEYAFDIKVWLTGTLSHQQTCLDGF 519
Query: 200 PEGELKKKMEMIFAESRQLLSNSLAVVSQVSQIVNAFQGGLSGFKLPWGKSDAHAPAPDA 259
+ M ++L S+ + S +++ L GF H
Sbjct: 520 VNTKTHAGETMA-----KVLKTSMELSSNAIDMMDVVSRILKGF---------HPSQYGV 657
Query: 260 DTDAVADDDEDLADAPDGAPDADQPIFEAPIGAPGAAPIGAPRVDAPPSWAAPAVD--LP 317
++DD PSW + L
Sbjct: 658 SRRLLSDD-------------------------------------GIPSWVSDGHRHLLA 726
Query: 318 GSTEKPTPNVTVAKDGSGDFKTISEALAAIPQTYKGRYVVYVKEGVYDETVTVTKKMVNL 377
G K N VA+DGSG FKT+++AL +P T +V+YVK GVY ETV V K+M +
Sbjct: 727 GGNVKA--NAVVAQDGSGQFKTLTDALKTVPPTNAAPFVIYVKAGVYKETVNVAKEMNYV 900
Query: 378 TMYGDGGLKSIITGNKNFVDGVRTFQTASFVVLGDGFVGRDMGFRNTAGAIKEQAVAARV 437
T+ GDG K+ TG+ N+ DG+ T++TA+F V G F+ +D+GF NTAG K QAVA RV
Sbjct: 901 TVIGDGPTKTKFTGSLNYADGINTYKTATFGVNGANFMAKDIGFENTAGTSKFQAVALRV 1080
Query: 438 QADCSIFVNCNFEGYQDTLYAQTHRQFYRDCVISGTIDFIFGHASAVFQNCQLVLRKPLD 497
AD +IF NC +G+QDTL+ ++ RQFYRDC ISGTIDF+FG A VFQNC+L+ R P
Sbjct: 1081TADQAIFHNCQMDGFQDTLFVESQRQFYRDCAISGTIDFVFGDAFGVFQNCKLICRVPAK 1260
Query: 498 NQKNIITANGRIDSKSNTAFVLQKCVIKGEDDLPSTTK--NYIGRPWKEYSRTIIMESDI 555
QK ++TA GR S +A V GE L S T +Y+GRPWK YS+ +IM+S I
Sbjct: 1261GQKCLVTAGGRDKQNSASALVFLSSHFTGEPALTSVTPKLSYLGRPWKLYSKVVIMDSTI 1440
Query: 556 PALIQPEGWLPWEGDFALKTLYYGEYDNVGAGAKTDARVKWIGRKDIKRGEALTYTVEPF 615
A+ PEG++P G T + EY+N G GA T+ RVKW G K + A Y F
Sbjct: 1441DAMFAPEGYMPMVGGAFKDTCTFYEYNNKGPGADTNLRVKWHGVKVLTSNVAAEYYPGKF 1620
Query: 616 LD-------GSWINGTGVPAHLG 631
+ +WI +GVP LG
Sbjct: 1621FEIVNATARDTWIVKSGVPYSLG 1689
>TC76733 similar to PIR|T06468|T06468 pectinesterase (EC 3.1.1.11) precursor
- garden pea, partial (98%)
Length = 2029
Score = 315 bits (806), Expect = 4e-86
Identities = 200/564 (35%), Positives = 296/564 (52%), Gaps = 14/564 (2%)
Frame = +2
Query: 83 CSHSEYKEKCVTTLKEALKKDPKLKEPKG----LLMVFMLVAKNEINNAFNKTANLK--F 136
C H+ + C+ + E + P L K +L+ + + I NA + + +K
Sbjct: 212 CEHAVDTKSCLAHVSE-VSHVPTLVTTKDQNLHVLLSLLTKSTTHIQNAMDTASVIKRRI 388
Query: 137 ASKEEKGAYEDCKQLFEDAKEEMGFSITEVGQLDISKLASKEAELNNWLSAVISYQDTCS 196
S E+ A DC+QL + + + ++ ++ + +I + + + WLS+V++ TC
Sbjct: 389 NSPREEIALSDCEQLMDLSMNRIWDTMLKLTKNNIDS----QQDAHTWLSSVLTNHATCL 556
Query: 197 DGFPEGELKKKMEMIFAESRQLLSNSLAVVSQVSQIVNAFQGGLSGFKLPWGKSDAHAPA 256
DG EG SR ++ N L +++ + L+ F + +
Sbjct: 557 DGL-EGS-----------SRVVMENDL------QDLISRARSSLAVFLVVF--------- 655
Query: 257 PDADTDAVADDDEDLADAPDGAPDADQPIFEAPIGAPGAAPIGAPRVDAPPSWAAPAVDL 316
P D D D+ + + P D+ + E +G A
Sbjct: 656 PQKDRDQFIDETL-IGEFPSWVTSKDRRLLETAVGDIKA--------------------- 769
Query: 317 PGSTEKPTPNVTVAKDGSGDFKTISEALAAIPQTYKGRYVVYVKEGVYDETVTVTKKMVN 376
NV VA+DGSG FKT++EA+A+ P K +YV+YVK+G Y E V + K N
Sbjct: 770 ---------NVVVAQDGSGKFKTVAEAVASAPDNGKTKYVIYVKKGTYKENVEIGSKKTN 922
Query: 377 LTMYGDGGLKSIITGNKNFVDGVRTFQTASFVVLGDGFVGRDMGFRNTAGAIKEQAVAAR 436
+ + GDG +IITGN NF+DG TF++++ +GDGF+ +D+ F+N AGA K QAVA R
Sbjct: 923 VMLVGDGMDATIITGNLNFIDGTTTFKSSTVAAVGDGFIAQDIWFQNMAGAAKHQAVALR 1102
Query: 437 VQADCSIFVNCNFEGYQDTLYAQTHRQFYRDCVISGTIDFIFGHASAVFQNCQLVLRKPL 496
V +D S+ C + +QDTLYA ++RQFYRD VI+GTIDFIFG+A+ VFQ C+LV RKP+
Sbjct: 1103VGSDQSVINRCRIDAFQDTLYAHSNRQFYRDSVITGTIDFIFGNAAVVFQKCKLVARKPM 1282
Query: 497 DNQKNIITANGRIDSKSNTAFVLQKCVIKGEDDLP---STTKNYIGRPWKEYSRTIIMES 553
NQ N+ TA GR D NT +Q+C + DL + K ++GRPWK+YSRT++M+S
Sbjct: 1283ANQNNMFTAQGREDPGQNTGTSIQQCDLTPSSDLKPVVGSIKTFLGRPWKKYSRTVVMQS 1462
Query: 554 DIPALIQPEGWLPWEG---DFALKTLYYGEYDNVGAGAKTDARVKWIGRKDIK-RGEALT 609
+ + I P GW W+ DF L+TLYYGEY N G GA T RV W G I EA
Sbjct: 1463FLDSHIDPTGWAEWDAASKDF-LQTLYYGEYLNNGPGAGTAKRVTWPGYHVINTAAEASK 1639
Query: 610 YTVEPFLDGS-WINGTGVPAHLGL 632
+TV + G+ W+ TGV GL
Sbjct: 1640FTVAQLIQGNVWLKNTGVAFTEGL 1711
>TC86403 similar to PIR|B86158|B86158 hypothetical protein AAF02886.1
[imported] - Arabidopsis thaliana, partial (63%)
Length = 1929
Score = 306 bits (784), Expect = 2e-83
Identities = 157/307 (51%), Positives = 202/307 (65%), Gaps = 7/307 (2%)
Frame = +3
Query: 327 VTVAKDGSGDFKTISEALAAIPQ---TYKGRYVVYVKEGVYDETVTVTKKMVNLTMYGDG 383
VTV++DGSG+F TI++A+AA P + G +++YV GVY+E +T+ KK L M GDG
Sbjct: 768 VTVSQDGSGNFTTINDAIAAAPNKSVSSDGYFLIYVTAGVYEEYITIDKKKTYLMMIGDG 947
Query: 384 GLKSIITGNKNFVDGVRTFQTASFVVLGDGFVGRDMGFRNTAGAIKEQAVAARVQADCSI 443
K+IITGN + VDG TF + +F V+G GFVG +M RNTAGA+K QAVA R AD S
Sbjct: 948 INKTIITGNHSVVDGWTTFGSPTFAVVGQGFVGVNMTIRNTAGAVKHQAVALRNGADLST 1127
Query: 444 FVNCNFEGYQDTLYAQTHRQFYRDCVISGTIDFIFGHASAVFQNCQLVLRKPLDNQKNII 503
F +C+FEGYQDTLY + RQFYR+C I GT+DFIFG+A VFQNC L R P+ Q N I
Sbjct: 1128 FYSCSFEGYQDTLYTHSLRQFYRECDIYGTVDFIFGNAKVVFQNCNLYPRLPMSGQFNAI 1307
Query: 504 TANGRIDSKSNTAFVLQKCVIKGEDDLPST---TKNYIGRPWKEYSRTIIMESDIPALIQ 560
TA GR D +T + C IK DDL ++ Y+GRPWKEYSRT+ M++ + +I
Sbjct: 1308 TAQGRTDPNQDTGTSIHNCTIKATDDLAASNGAVSTYLGRPWKEYSRTVYMQTFMDNVIN 1487
Query: 561 PEGWLPWEGDFALKTLYYGEYDNVGAGAKTDARVKWIGRKDIKRGEALTYTVEPFLDG-S 619
GW W+G+FAL TLYY E++N G G+ TD RV W G I +A +TV FL G
Sbjct: 1488 VAGWRAWDGEFALSTLYYAEFNNSGPGSSTDGRVTWQGYHVINATDAANFTVANFLLGDD 1667
Query: 620 WINGTGV 626
W+ TGV
Sbjct: 1668 WLPQTGV 1688
>TC85623 similar to GP|13605696|gb|AAK32841.1 At2g45220/F4L23.27 {Arabidopsis
thaliana}, partial (79%)
Length = 2442
Score = 300 bits (767), Expect = 1e-81
Identities = 158/334 (47%), Positives = 208/334 (61%), Gaps = 5/334 (1%)
Frame = +3
Query: 304 DAPPSWAAPAVD--LPGSTEKPTPNVTVAKDGSGDFKTISEALAAIPQTYKGRYVVYVKE 361
D P+W P L S+ NV VAKDGSG + T+ A A P GRYV+YVK
Sbjct: 1104 DGFPTWVKPGDRKLLQTSSAASKANVVVAKDGSGKYTTVKAATDAAPSG-SGRYVIYVKA 1280
Query: 362 GVYDETVTVTKKMVNLTMYGDGGLKSIITGNKNFVDGVRTFQTASFVVLGDGFVGRDMGF 421
GVY+E V + K N+ + GDG K+IITG+K+ G TF++A+ GDGF+ +D+ F
Sbjct: 1281 GVYNEQVEIKAK--NVMLVGDGIGKTIITGSKSVGGGTTTFRSATVAATGDGFIAQDITF 1454
Query: 422 RNTAGAIKEQAVAARVQADCSIFVNCNFEGYQDTLYAQTHRQFYRDCVISGTIDFIFGHA 481
RNTAGA QAVA R +D S+F C+FEGYQDTLY + RQFYR+C I GT+DFIFG+A
Sbjct: 1455 RNTAGAANHQAVAFRSGSDLSVFYKCSFEGYQDTLYVHSERQFYRECNIYGTVDFIFGNA 1634
Query: 482 SAVFQNCQLVLRKPLDNQKNIITANGRIDSKSNTAFVLQKCVIKGEDDL-PSTTKNYIGR 540
+ V QNC + R P + +TA GR D NT ++ + + DL PS+ K+Y+GR
Sbjct: 1635 AVVLQNCNIFARNP-PAKTITVTAQGRTDPNQNTGIIIHNSRVSAQSDLNPSSVKSYLGR 1811
Query: 541 PWKEYSRTIIMESDIPALIQPEGWLPWEGDFALKTLYYGEYDNVGAGAKTDARVKWIGRK 600
PW++YSRT+ M++ + I P GWLPW+G+FAL TLYY EY N G+G+ T RV W G
Sbjct: 1812 PWQKYSRTVFMKTVLDGFINPAGWLPWDGNFALDTLYYAEYANTGSGSSTSNRVTWKGYH 1991
Query: 601 DI-KRGEALTYTVEPFLDG-SWINGTGVPAHLGL 632
+ +A +TV F+ G SWI TGVP GL
Sbjct: 1992 VLTSASQASPFTVGNFIAGNSWIGNTGVPFTSGL 2093
>TC77013 similar to GP|7025485|gb|AAF35897.1| pectin methylesterase isoform
alpha {Vigna radiata}, partial (94%)
Length = 1655
Score = 295 bits (755), Expect(2) = 4e-80
Identities = 149/297 (50%), Positives = 192/297 (64%), Gaps = 4/297 (1%)
Frame = +2
Query: 326 NVTVAKDGSGDFKTISEALAAIPQTYKGRYVVYVKEGVYDETVTVTKKMVNLTMYGDGGL 385
++ VAKDG+G+F TI EA+AA P + R+V+++K G Y E V V KK NL + GDG
Sbjct: 98 DLVVAKDGTGNFATIGEAIAAAPNSSATRFVIHIKAGAYFENVEVIKKKTNLMLVGDGIG 277
Query: 386 KSIITGNKNFVDGVRTFQTASFVVLGDGFVGRDMGFRNTAGAIKEQAVAARVQADCSIFV 445
++++ ++N VDG TFQ+++F V+GD F+ + + F N+AG K QAVA R AD S F
Sbjct: 278 QTVVKASRNVVDGWTTFQSSTFAVVGDKFIAKGITFENSAGPSKHQAVAVRNGADFSAFY 457
Query: 446 NCNFEGYQDTLYAQTHRQFYRDCVISGTIDFIFGHASAVFQNCQLVLRKPLDNQKNIITA 505
C+F YQDTLY + RQFYR+C + GT+DFIFG+A+ VFQNC L RKP QKN+ TA
Sbjct: 458 QCSFVAYQDTLYVHSLRQFYRECDVYGTVDFIFGNAAVVFQNCNLYARKPDPKQKNLFTA 637
Query: 506 NGRIDSKSNTAFVLQKCVIKGEDDL---PSTTKNYIGRPWKEYSRTIIMESDIPALIQPE 562
GR D NT + C I DL ST K+Y+GRPWK+YSRT+ + S I LI P
Sbjct: 638 QGREDPNQNTGISILNCKIAAAADLIPVQSTFKSYLGRPWKKYSRTVYLNSFIDNLIDPP 817
Query: 563 GWLPWEGDFALKTLYYGEYDNVGAGAKTDARVKWIGRKDIKRG-EALTYTVEPFLDG 618
GWL W G FAL TL+YGEY N G G+ T ARV W G K I EA +TV F+ G
Sbjct: 818 GWLEWNGTFALDTLFYGEYKNRGPGSNTSARVTWPGYKVITNATEASQFTVRQFIQG 988
Score = 21.9 bits (45), Expect(2) = 4e-80
Identities = 8/13 (61%), Positives = 9/13 (68%)
Frame = +1
Query: 620 WINGTGVPAHLGL 632
W+N TGVP L L
Sbjct: 997 WLNSTGVPFFLDL 1035
>TC77735 similar to GP|14334646|gb|AAK59501.1 putative pectinesterase
{Arabidopsis thaliana}, partial (59%)
Length = 1963
Score = 293 bits (750), Expect = 1e-79
Identities = 150/315 (47%), Positives = 198/315 (62%), Gaps = 7/315 (2%)
Frame = +2
Query: 327 VTVAKDGSGDFKTISEALAAIPQTYK---GRYVVYVKEGVYDETVTVTKKMVNLTMYGDG 383
V V+ G + +I +A+AA P K G Y++YV+EG Y+E V V K N+ + GDG
Sbjct: 827 VLVSPYGIANHTSIGDAIAAAPNNTKPEDGYYLIYVREGYYEEYVIVPKHKNNILLVGDG 1006
Query: 384 GLKSIITGNKNFVDGVRTFQTASFVVLGDGFVGRDMGFRNTAGAIKEQAVAARVQADCSI 443
+IITGN + +DG TF +++F V G+ F+ D+ FRNTAG K QAVA R AD S
Sbjct: 1007 INNTIITGNHSVIDGWTTFNSSTFAVSGERFIAVDITFRNTAGPEKHQAVAVRNNADLST 1186
Query: 444 FVNCNFEGYQDTLYAQTHRQFYRDCVISGTIDFIFGHASAVFQNCQLVLRKPLDNQKNII 503
F C+FEGYQDTLY + RQFYRDC I GT+DFIFG+A+ VFQNC + RKPL NQKN +
Sbjct: 1187 FYRCSFEGYQDTLYVHSVRQFYRDCKIYGTVDFIFGNAAVVFQNCNIYARKPLPNQKNAV 1366
Query: 504 TANGRIDSKSNTAFVLQKCVIKGEDDLP---STTKNYIGRPWKEYSRTIIMESDIPALIQ 560
TA GR D NT +Q C I DL ++T +Y+GRPWK YSRT+ M+S I +Q
Sbjct: 1367 TAQGRTDPNQNTGISIQNCTIDAAQDLANDLNSTMSYLGRPWKIYSRTVYMQSYIGDFVQ 1546
Query: 561 PEGWLPWEGDFALKTLYYGEYDNVGAGAKTDARVKWIGRKDIKRGEALTYTVEPF-LDGS 619
P GWL W G L T++YGE++N G G+ T+ RV+W G + +A +TV F L +
Sbjct: 1547 PSGWLEWNGTVGLDTIFYGEFNNYGPGSVTNNRVQWPGHFLLNDTQAWNFTVLNFTLGNT 1726
Query: 620 WINGTGVPAHLGLYN 634
W+ T +P GL N
Sbjct: 1727 WLPDTDIPYTEGLLN 1771
Score = 32.7 bits (73), Expect = 0.44
Identities = 39/176 (22%), Positives = 68/176 (38%), Gaps = 10/176 (5%)
Frame = +2
Query: 63 QNAPPEPERVDKYSRLVTMLCSHSEYKEKCVTTLKEALKKDPKLKEPKGLLMVFM-LVAK 121
Q+ PP P S + C + Y + C + L A++ P G + L
Sbjct: 101 QSPPPSPSPPPPSSS--SAACKTTLYPKLCRSMLS-AIRSSPSDPYNYGKFSIKQNLKVA 271
Query: 122 NEINNAF----NKTANLKFASKEEKGAYEDCKQLFE---DAKEEMGFSITEVGQLDISKL 174
++ F N+ + + EE GA DCK L D E + + S
Sbjct: 272 RKLEKVFIDFLNRHQSSSSLNHEEVGALVDCKDLNSLNVDYLESISDELKSASSSSSSSD 451
Query: 175 ASKEAELNNWLSAVISYQDTCSDGF--PEGELKKKMEMIFAESRQLLSNSLAVVSQ 228
++ ++LSAV + TC DG + + + + ++ Q S SL +V++
Sbjct: 452 TELVDKIESYLSAVATNHYTCYDGLVVTKSNIANALAVPLKDATQFYSVSLGLVTE 619
>BI272853 weakly similar to PIR|T04359|T04 pectin methylesterase-like protein
- maize, partial (25%)
Length = 678
Score = 213 bits (542), Expect = 2e-55
Identities = 108/206 (52%), Positives = 139/206 (67%), Gaps = 3/206 (1%)
Frame = +1
Query: 353 GRYVVYVKEGVYDETVTVTKKMVNLTMYGDGGLKSIITGNKNFVDGVRTFQTASFVVLGD 412
GRY +YVK GVYDE +T+ K VN+ MYGDG K+I+TG KN GV+T QTA+F
Sbjct: 1 GRYTIYVKAGVYDEYITIPKDAVNILMYGDGPGKTIVTGRKNGAAGVKTMQTATFANTAL 180
Query: 413 GFVGRDMGFRNTAGAIKEQAVAARVQADCSIFVNCNFEGYQDTLYAQTHRQFYRDCVISG 472
GF+G+ M F NTAG QAVA R Q D S V C+ GYQDTLY QT+RQFYR+CVISG
Sbjct: 181 GFIGKAMTFENTAGPAGHQAVAFRNQGDMSALVGCHILGYQDTLYVQTNRQFYRNCVISG 360
Query: 473 TIDFIFGHASAVFQNCQLVLRKPLDNQKNIITANGRIDSKSNTAFVLQKCVIKGEDDL-- 530
T+DFIFG ++ + Q+ +++R P NQ N ITA+G +K NT V+Q C I + L
Sbjct: 361 TVDFIFGTSATLIQDSTIIVRMPSPNQFNTITADGSYVNKLNTGIVIQGCNIVPKAALFP 540
Query: 531 -PSTTKNYIGRPWKEYSRTIIMESDI 555
T K+Y+GRPWK ++T++MES I
Sbjct: 541 QRFTIKSYLGRPWKVLTKTVVMESTI 618
>TC84419 similar to SP|Q42920|PME_MEDSA Pectinesterase precursor (EC
3.1.1.11) (Pectin methylesterase) (PE) (P65). [Alfalfa],
partial (53%)
Length = 732
Score = 209 bits (533), Expect = 2e-54
Identities = 101/199 (50%), Positives = 134/199 (66%), Gaps = 2/199 (1%)
Frame = +2
Query: 414 FVGRDMGFRNTAGAIKEQAVAARVQADCSIFVNCNFEGYQDTLYAQTHRQFYRDCVISGT 473
F ++GF N+AGA K QAVA RV AD ++F NC GYQDTLY Q+ RQFYRDC I+GT
Sbjct: 86 FTALNVGFENSAGAAKHQAVALRVTADKALFYNCQMNGYQDTLYTQSKRQFYRDCTITGT 265
Query: 474 IDFIFGHASAVFQNCQLVLRKPLDNQKNIITANGRIDSKSNTAFVLQKCVIKGEDDLPST 533
IDF+FG A VFQNC+L++RKP+ NQ+ ++TA GR S +A V Q C GE ++ +
Sbjct: 266 IDFVFGDAVGVFQNCKLIVRKPMANQQCMVTAGGRTKVDSVSALVFQNCHFTGEPEVLTM 445
Query: 534 TKN--YIGRPWKEYSRTIIMESDIPALIQPEGWLPWEGDFALKTLYYGEYDNVGAGAKTD 591
Y+GRPW+ +S+ +I++S I L PEG++PW G+ +T Y EY+N GAGA T+
Sbjct: 446 QPKIAYLGRPWRNFSKVVIVDSLIDGLFVPEGYMPWMGNLFKETCTYLEYNNKGAGAATN 625
Query: 592 ARVKWIGRKDIKRGEALTY 610
+VKW G K I GEA Y
Sbjct: 626 LKVKWPGVKTISAGEAAKY 682
>TC91752 similar to GP|6630559|gb|AAF19578.1| putative pectinesterase
{Arabidopsis thaliana}, partial (34%)
Length = 691
Score = 203 bits (517), Expect = 1e-52
Identities = 97/194 (50%), Positives = 132/194 (68%)
Frame = +3
Query: 322 KPTPNVTVAKDGSGDFKTISEALAAIPQTYKGRYVVYVKEGVYDETVTVTKKMVNLTMYG 381
K ++ VAKDGSG +KTIS AL +P R V+YVK+G+Y E V V K N+ + G
Sbjct: 108 KKKADIVVAKDGSGKYKTISAALKHVPNKSDKRTVIYVKKGIYYENVRVEKTKWNVMIIG 287
Query: 382 DGGLKSIITGNKNFVDGVRTFQTASFVVLGDGFVGRDMGFRNTAGAIKEQAVAARVQADC 441
DG +I++G+ NF+DG TF TA+F V G F+ RD+GF+NTAG K QAVA AD
Sbjct: 288 DGMNVTIVSGSLNFIDGTPTFSTATFAVFGRNFIARDIGFKNTAGPQKHQAVALMTSADQ 467
Query: 442 SIFVNCNFEGYQDTLYAQTHRQFYRDCVISGTIDFIFGHASAVFQNCQLVLRKPLDNQKN 501
++F C+ + YQDTLYA ++RQFYR+C I GT+DFIFG+++ V QNC ++ R+P+ Q+N
Sbjct: 468 AVFYKCSMDAYQDTLYAHSNRQFYRECNIYGTVDFIFGNSAVVLQNCNILPRQPMPGQQN 647
Query: 502 IITANGRIDSKSNT 515
ITA G+ D NT
Sbjct: 648 TITAQGKTDPNMNT 689
>BQ138825 similar to PIR|T49922|T49 pectin methylesterase-like protein -
Arabidopsis thaliana, partial (36%)
Length = 658
Score = 198 bits (503), Expect = 6e-51
Identities = 103/218 (47%), Positives = 140/218 (63%), Gaps = 14/218 (6%)
Frame = +1
Query: 342 EALAAIPQTY--KGRYVVYVKEGVYDETVTVTKKMVNLTMYGDGGLKSIITGNKNFVD-G 398
EA+ A P + R+V+Y+KEGVY+ETV V K N+ GDG K++ITG+ N G
Sbjct: 4 EAVNAAPDNGVDRKRFVIYIKEGVYEETVRVPLKKRNVVFLGDGIGKTVITGSANVGQPG 183
Query: 399 VRTFQTASFVVLGDGFVGRDMGFRNTAGAIKEQAVAARVQADCSIFVNCNFEGYQDTLYA 458
+ T+ +A+ VLGDGF+ +D+ NTAG QAVA R+ +D S+ NC F G QDTLYA
Sbjct: 184 MTTYNSATVAVLGDGFMAKDLTIENTAGPDAHQAVAFRLDSDLSVIENCEFLGNQDTLYA 363
Query: 459 QTHRQFYRDCVISGTIDFIFGHASAVFQNCQLVLR----KPLDNQKNIITANGRIDSKSN 514
+ RQFY+ C I G +DFIFG+++A+FQ+CQ+++R KP + N ITA+GR D +
Sbjct: 364 HSLRQFYKSCRIVGNVDFIFGNSAAIFQDCQILVRPRQLKPEKGENNAITAHGRTDPAQS 543
Query: 515 TAFVLQKCVIKGEDDL-------PSTTKNYIGRPWKEY 545
T FV Q C+I G +D P KNY+GRPWKEY
Sbjct: 544 TGFVFQNCLINGTEDYMALYHSNPKVHKNYLGRPWKEY 657
>BE319906 similar to SP|Q43062|PME_ Pectinesterase PPE8B precursor (EC
3.1.1.11) (Pectin methylesterase) (PE). [Peach] {Prunus
persica}, partial (36%)
Length = 589
Score = 185 bits (470), Expect = 4e-47
Identities = 86/162 (53%), Positives = 120/162 (73%)
Frame = +2
Query: 333 GSGDFKTISEALAAIPQTYKGRYVVYVKEGVYDETVTVTKKMVNLTMYGDGGLKSIITGN 392
GSGD+ + +A++A P++ RYV+YVK+GVY E V + KK N+ + G+G +II+G+
Sbjct: 2 GSGDYAKVMDAVSAAPESSMKRYVIYVKKGVYVENVEIKKKKWNIMLIGEGMDATIISGS 181
Query: 393 KNFVDGVRTFQTASFVVLGDGFVGRDMGFRNTAGAIKEQAVAARVQADCSIFVNCNFEGY 452
+N+VDG TF++A+F V G GF+ RD+ F+NTAGA K QAVA R +D S+F C GY
Sbjct: 182 RNYVDGSTTFRSATFAVSGRGFIARDISFQNTAGAEKHQAVALRSDSDLSVFYRCGIFGY 361
Query: 453 QDTLYAQTHRQFYRDCVISGTIDFIFGHASAVFQNCQLVLRK 494
QD+LY T RQFYR+C ISGT+DFIFG A+AVFQNCQ++ ++
Sbjct: 362 QDSLYTHTMRQFYRECKISGTVDFIFGDATAVFQNCQILAKE 487
>BG455363 weakly similar to PIR|T52331|T523 pectinesterase (EC 3.1.1.11)
[imported] - Salix gilgiana, partial (7%)
Length = 685
Score = 155 bits (393), Expect = 3e-38
Identities = 76/189 (40%), Positives = 122/189 (64%), Gaps = 2/189 (1%)
Frame = +3
Query: 1 MAFEDFDLVSERRKANAKQHLKKKILIGVTSVVLIACVIAAVTFVIVKRSGPDHNNNDKK 60
MAF+DFDL++ERRK+ + ++K+I+IGV S V+ +I FV + P
Sbjct: 117 MAFQDFDLINERRKSEKRAQMRKRIIIGVISSVVFVGLIGCAFFVATTKYNPFGGGGGGS 296
Query: 61 PVQNAPPE--PERVDKYSRLVTMLCSHSEYKEKCVTTLKEALKKDPKLKEPKGLLMVFML 118
+++ + V ++V ++CS ++YKEKC L +A++KDPKL+ PK LL V++
Sbjct: 297 HPEDSSTANGSKHVAHSEKVVKLVCSSADYKEKCEGPLNKAVEKDPKLQHPKDLLKVYLK 476
Query: 119 VAKNEINNAFNKTANLKFASKEEKGAYEDCKQLFEDAKEEMGFSITEVGQLDISKLASKE 178
++E+N AFNKT + KF +KEEK A+EDCK++ + AK+++ SI ++ + D+ KLASK
Sbjct: 477 TVEDEVNKAFNKTNSFKFNTKEEKAAFEDCKEMIQYAKDDLATSIDQLSEADMKKLASKT 656
Query: 179 AELNNWLSA 187
+LN+W SA
Sbjct: 657 PDLNSWXSA 683
>AL368659 similar to PIR|B86158|B86 hypothetical protein AAF02886.1
[imported] - Arabidopsis thaliana, partial (25%)
Length = 495
Score = 149 bits (376), Expect = 3e-36
Identities = 75/145 (51%), Positives = 101/145 (68%), Gaps = 3/145 (2%)
Frame = +1
Query: 331 KDGSGDFKTISEALAAIPQTY---KGRYVVYVKEGVYDETVTVTKKMVNLTMYGDGGLKS 387
+DGSG+F TI++A+AA P G +++++ EGVY+E V++ KK L M G+G ++
Sbjct: 61 QDGSGNFTTINDAIAAAPNNTVASDGYFLIFITEGVYEEYVSIDKKKKYLMMIGEGINQT 240
Query: 388 IITGNKNFVDGVRTFQTASFVVLGDGFVGRDMGFRNTAGAIKEQAVAARVQADCSIFVNC 447
IITGN+N DG TF +A+F V+ GFV ++ FRNTAGA K QAVA R AD S F +C
Sbjct: 241 IITGNRNVADGFTTFNSATFAVVAQGFVAVNITFRNTAGAAKNQAVALRSGADMSTFYSC 420
Query: 448 NFEGYQDTLYAQTHRQFYRDCVISG 472
+FEGYQDTLY + RQFYR+C I G
Sbjct: 421 SFEGYQDTLYTHSLRQFYRECDIYG 495
>TC89627 weakly similar to GP|17529290|gb|AAL38872.1 putative pectin
methylesterase {Arabidopsis thaliana}, partial (33%)
Length = 1289
Score = 119 bits (297), Expect(2) = 2e-33
Identities = 73/224 (32%), Positives = 114/224 (50%), Gaps = 10/224 (4%)
Frame = +3
Query: 329 VAKDGSGDFKTISEALAAIPQTYKGRYVVYVKEGVYDETVTVTKKMVNLTMYGDGGLKSI 388
V++DGS FK+I+EAL +I R ++ + G Y E + V K + +T GD
Sbjct: 243 VSQDGSAQFKSITEALNSIQPYNIRRVIISIAPGYYREKIVVPKTLPFITFLGDVRDPPT 422
Query: 389 ITGNKNFV----DG--VRTFQTASFVVLGDGFVGRDMGFRNTA----GAIKEQAVAARVQ 438
ITGN DG +RTF +A+ V F+ ++ F NTA G+ EQAVA R+
Sbjct: 423 ITGNDTQSVTGSDGAQLRTFNSATVAVNASYFMAININFENTASFPIGSKVEQAVAVRIT 602
Query: 439 ADCSIFVNCNFEGYQDTLYAQTHRQFYRDCVISGTIDFIFGHASAVFQNCQLVLRKPLDN 498
+ + F NC F G QDTLY ++ +C I G++DFI GH ++++ C + + + N
Sbjct: 603 GNKTAFYNCTFSGVQDTLYDHKGLHYFNNCTIKGSVDFICGHGKSLYEGCTI---RSIAN 773
Query: 499 QKNIITANGRIDSKSNTAFVLQKCVIKGEDDLPSTTKNYIGRPW 542
ITA + ++ F + ++ G+ Y+GRPW
Sbjct: 774 NMTSITAQSGSNPSYDSGFSFKNSMVIGDG------PTYLGRPW 887
Score = 42.0 bits (97), Expect(2) = 2e-33
Identities = 23/78 (29%), Positives = 36/78 (45%), Gaps = 1/78 (1%)
Frame = +1
Query: 545 YSRTIIMESDIPALIQPEGWLPWEGDFALKTLYYGEYDNVGAGAKTDARVKWIGRKDIKR 604
YS+ + + + + P+GW W YYGEY G G+ T RV W + K
Sbjct: 895 YSQVVFSYTYMDNSVLPKGWEDWNDTKRYMNAYYGEYKCSGPGSNTAGRVPWARMLNDK- 1071
Query: 605 GEALTYTVEPFLDG-SWI 621
EA + ++DG +W+
Sbjct: 1072-EAQVFIGTQYIDGNTWL 1122
>TC92167 similar to PIR|T05203|T05203 pectinesterase homolog F4I10.160 -
Arabidopsis thaliana, partial (11%)
Length = 652
Score = 115 bits (287), Expect(2) = 2e-28
Identities = 70/186 (37%), Positives = 102/186 (54%), Gaps = 2/186 (1%)
Frame = +3
Query: 359 VKEGVYDETVTVTKKMVNLTMYGDGGLKSIITGNKNFVDGVRTFQTASFVVLGDGFVGRD 418
+K+ DE VT+T KM ++T+YGDG KSIITG+KNF DGV TAS VVLG+GF+G
Sbjct: 33 LKKECTDEVVTITDKMKDITIYGDGSQKSIITGSKNFRDGVTNINTASLVVLGEGFLGLA 212
Query: 419 MGFRNTAGAIKEQAVAA-RVQADCSIFVNCNFEGYQDTLYAQTHRQFYRDCVISGTIDF- 476
G + + + + + ++ C + + +++ T GTI F
Sbjct: 213 SGIQKHSWS*RTSSSSS*SPSRSCCVCQLPF*RLPRHAIHSGT*TILPVAAS*QGTIRFH 392
Query: 477 IFGHASAVFQNCQLVLRKPLDNQKNIITANGRIDSKSNTAFVLQKCVIKGEDDLPSTTKN 536
++ +FQNC LV++KP Q N +TA GR+D+K NTA VL KC +K +D L +
Sbjct: 393 LW*PQPVIFQNCILVVKKPSVGQSNAVTAQGRLDNKQNTAIVLHKCTLKADDALVPVKSH 572
Query: 537 YIGRPW 542
PW
Sbjct: 573 SKKLPW 590
Score = 95.9 bits (237), Expect = 4e-20
Identities = 61/145 (42%), Positives = 79/145 (54%), Gaps = 8/145 (5%)
Frame = +1
Query: 419 MGFRNTAGAIKEQAVAARVQADCSIFVNCNFEGYQDTLYAQTHRQFYRDCVIS-GTIDFI 477
+GFRNTAG QAVAARVQAD ++F NC FEG+QDTLY HRQF+ + DFI
Sbjct: 214 VGFRNTAGPEGHQAVAARVQADRAVFANCRFEGFQDTLYTVAHRQFFP*LHHNREQFDFI 393
Query: 478 FGHASAVFQNCQLVLRKPLDNQKNIITANGRIDSKSNTAFVLQKCVIKGEDD-------L 530
FG S ++ L L NQ + + T + Q I L
Sbjct: 394 FGDPSPLYSKTASWL---LKNQVSAKVTR*QHKEDWTTNKIQQLFYINVHLKLMTHWFLL 564
Query: 531 PSTTKNYIGRPWKEYSRTIIMESDI 555
+T K+Y+GR WK++SRT++MESDI
Sbjct: 565 KATVKSYLGRAWKQFSRTVVMESDI 639
Score = 29.3 bits (64), Expect(2) = 2e-28
Identities = 11/14 (78%), Positives = 13/14 (92%)
Frame = +2
Query: 351 YKGRYVVYVKEGVY 364
+ GRYV+YVKEGVY
Sbjct: 8 FLGRYVIYVKEGVY 49
>TC82059 similar to GP|19424045|gb|AAL87311.1 unknown protein {Arabidopsis
thaliana}, partial (17%)
Length = 483
Score = 113 bits (283), Expect = 2e-25
Identities = 55/102 (53%), Positives = 65/102 (62%), Gaps = 1/102 (0%)
Frame = +1
Query: 532 STTKNYIGRPWKEYSRTIIMESDIPALIQPEGWLPWEGDFALKTLYYGEYDNVGAGAKTD 591
+T K Y+GRPWKEYSRT+ M+S + + I P GW W GDFAL TLYY EYDN GAG+ T
Sbjct: 7 ATVKTYLGRPWKEYSRTVYMQSFMDSFINPSGWREWNGDFALSTLYYAEYDNRGAGSSTA 186
Query: 592 ARVKWIGRKDIKRGEALTYTVEPFLDG-SWINGTGVPAHLGL 632
RV W G I +A +TV F+ G WI TGVP GL
Sbjct: 187 NRVTWPGYHVIGATDAANFTVSNFVSGDDWIPQTGVPYMSGL 312
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.316 0.134 0.395
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 17,044,143
Number of Sequences: 36976
Number of extensions: 224407
Number of successful extensions: 1282
Number of sequences better than 10.0: 87
Number of HSP's better than 10.0 without gapping: 1201
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1237
length of query: 634
length of database: 9,014,727
effective HSP length: 102
effective length of query: 532
effective length of database: 5,243,175
effective search space: 2789369100
effective search space used: 2789369100
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)
S2: 61 (28.1 bits)
Medicago: description of AC130802.4


