

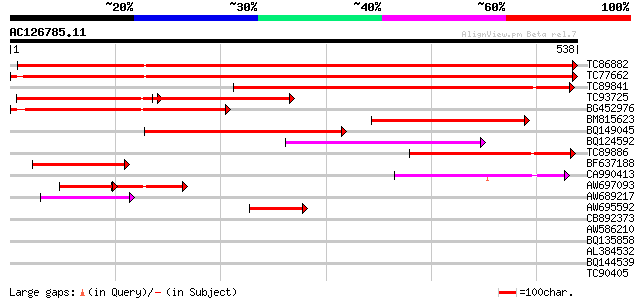
BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC126785.11 + phase: 0
(538 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC86882 similar to GP|10177467|dbj|BAB10858. permease 1 {Arabido... 847 0.0
TC77662 similar to GP|10177467|dbj|BAB10858. permease 1 {Arabido... 840 0.0
TC89841 similar to PIR|T02307|T02307 probable membrane transport... 397 e-111
TC93725 similar to GP|10177467|dbj|BAB10858. permease 1 {Arabido... 218 6e-57
BG452976 weakly similar to GP|15983805|gb At1g49960/F2J10_14 {Ar... 212 3e-55
BM815623 similar to GP|14334908|gb putative membrane transporter... 200 1e-51
BQ149045 similar to PIR|T02307|T02 probable membrane transporter... 177 1e-44
BQ124592 similar to GP|20466159|gb putative membrane transporter... 154 1e-37
TC89886 weakly similar to GP|14587294|dbj|BAB61205. putative per... 151 7e-37
BF637188 similar to GP|14334908|gb putative membrane transporter... 118 7e-27
CA990413 similar to PIR|T05632|T05 hypothetical protein F20D10.1... 105 6e-23
AW697093 similar to GP|20466159|gb putative membrane transporter... 56 5e-17
AW689217 similar to GP|15144504|gb| putative permease {Lycopersi... 65 9e-11
AW695592 weakly similar to PIR|T00984|T009 probable membrane tra... 61 1e-09
CB892373 PIR|D64842|D64 probable transport protein ycdG - Escher... 39 0.005
AW586210 33 0.36
BQ135858 similar to GP|23491723|db formin homology protein A {Di... 32 0.47
AL384532 32 0.80
BQ144539 GP|13173388|g lysine/glutamic acid-rich protein {Candid... 30 1.8
TC90405 similar to GP|18086395|gb|AAL57656.1 AT5g13210/T31B5_30 ... 30 3.1
>TC86882 similar to GP|10177467|dbj|BAB10858. permease 1 {Arabidopsis
thaliana}, complete
Length = 2140
Score = 847 bits (2187), Expect = 0.0
Identities = 409/531 (77%), Positives = 465/531 (87%)
Frame = +2
Query: 8 GGATPPPKQDEFQPHPVKDQLPNVSYCITSPPPWPEAIMLGFQHYLVMLGTTVLIPTALV 67
GG P PK DE QPHP KDQLPNVSYCITSPPPWPEAI+LGFQHYLVMLGTTVLIPT+LV
Sbjct: 281 GGGAPAPKADEPQPHPPKDQLPNVSYCITSPPPWPEAILLGFQHYLVMLGTTVLIPTSLV 460
Query: 68 SQMGGGNEEKAMLIQNHLFVAGINTLIQTLFGTRLPAVIGGSFTFVPTTISIILASRYDD 127
QMGGGNEEKA +IQ LFVAGINTL+QTLFG+RLPAVIGGS+TFVP TISIILA R++D
Sbjct: 461 PQMGGGNEEKAKVIQTLLFVAGINTLVQTLFGSRLPAVIGGSYTFVPATISIILAGRFND 640
Query: 128 DIMHPREKFKRIMRGTQGALIVASSLQIIVGFSGLWCHVVRFISPLSAVPLVALTGFGLY 187
+ P EK K+IMR TQGALIVAS+LQI++GFSGLW +V RF+SPLSAVPLV+L GFGLY
Sbjct: 641 E-PDPIEKIKKIMRATQGALIVASTLQIVLGFSGLWRNVARFLSPLSAVPLVSLVGFGLY 817
Query: 188 ELGFPMLAKCIEIGLPEIVILVFLSQFMPHMMKGGRHIFARFAVIFSVIIVWVYAIILTG 247
ELGFP +AKC+EIGLPE+V+LVF+SQF+PH++ G+H+F RF+V+F+V IVW+YA ILT
Sbjct: 818 ELGFPGVAKCVEIGLPELVLLVFVSQFVPHVLHSGKHVFDRFSVLFTVAIVWLYAFILTV 997
Query: 248 CGAYKNAEHETQDTCRTDRAGLIHGASWISPPIPFRWGAPTFDAGEAFAMMAASFVAQIE 307
GAY + + TQ TCRTD +GLI A WI P PF+WGAP+FDAGEAFAMM SFVA +E
Sbjct: 998 GGAYNHVKRTTQMTCRTDSSGLIDAAPWIRVPYPFQWGAPSFDAGEAFAMMMTSFVALVE 1177
Query: 308 STGGFIAVARFASATPVPPSVLSRGIGWQGVGILLSGIFGTGNGSSVSIENAGLLALTRV 367
S+G FIAV RFASATP+PPS+LSRGIGWQGVGILLSG+FGTG GSSVS+ENAGLLALTRV
Sbjct: 1178SSGAFIAVYRFASATPLPPSILSRGIGWQGVGILLSGLFGTGIGSSVSVENAGLLALTRV 1357
Query: 368 GSRRVVQISAGFMIFFSILGKFGAVFASIPMPIVAALYCLLFSQVGSAGLSFLQFCNLNS 427
GSRRVVQISAGFMIFFSILGKFGAVFASIP PIVAALYCL F+ VGS GLSFLQFCNLNS
Sbjct: 1358GSRRVVQISAGFMIFFSILGKFGAVFASIPPPIVAALYCLFFAYVGSGGLSFLQFCNLNS 1537
Query: 428 FRTKFIIGFSIFMGFSVPQYFKEYTAIKQYGPVHTNARWFNDMINVPFSSGAFVAGILAL 487
FRTKF++GFSIF+G S+PQYF EYTAI +GPVHT ARWFND++NVPF S AFVAG++A
Sbjct: 1538FRTKFVLGFSIFLGLSIPQYFNEYTAINGFGPVHTGARWFNDIVNVPFQSKAFVAGVVAY 1717
Query: 488 FFDVTLHKSDNQTRKDRGMHWWDRFSSFKTDTRSEEFYSLPFNLNKFFPSV 538
F D TLHK ++ RKDRG HWWD++ SFKTDTRSEEFYSLPFNLNK+FPSV
Sbjct: 1718FLDNTLHKKESAIRKDRGKHWWDKYRSFKTDTRSEEFYSLPFNLNKYFPSV 1870
>TC77662 similar to GP|10177467|dbj|BAB10858. permease 1 {Arabidopsis
thaliana}, complete
Length = 1953
Score = 840 bits (2169), Expect = 0.0
Identities = 407/538 (75%), Positives = 468/538 (86%)
Frame = +2
Query: 1 MAGGGGGGGATPPPKQDEFQPHPVKDQLPNVSYCITSPPPWPEAIMLGFQHYLVMLGTTV 60
MAGGGGG P K DE QPHP KDQLPN+SYCITSPPPWPEAI+LGFQH+LVMLGTTV
Sbjct: 101 MAGGGGG-----PAKSDEPQPHPPKDQLPNISYCITSPPPWPEAILLGFQHFLVMLGTTV 265
Query: 61 LIPTALVSQMGGGNEEKAMLIQNHLFVAGINTLIQTLFGTRLPAVIGGSFTFVPTTISII 120
LIPTALV QMGGGN EKA +I+ LFVAGINTL+QTLFG+RLPAVIGGS+T+VPTTISII
Sbjct: 266 LIPTALVPQMGGGNAEKAKVIETLLFVAGINTLVQTLFGSRLPAVIGGSYTYVPTTISII 445
Query: 121 LASRYDDDIMHPREKFKRIMRGTQGALIVASSLQIIVGFSGLWCHVVRFISPLSAVPLVA 180
LA R+ ++ P EKFK+IMR QGALIVAS+LQI++GFSGLW +V RF+SPLSAVPLV+
Sbjct: 446 LAGRFSNE-PDPIEKFKKIMRAVQGALIVASTLQIVLGFSGLWRNVARFLSPLSAVPLVS 622
Query: 181 LTGFGLYELGFPMLAKCIEIGLPEIVILVFLSQFMPHMMKGGRHIFARFAVIFSVIIVWV 240
L GFGLYELGFP +AKC+EIGLPE+++LVF+SQ++PH++ G++IF RF+V+F++ IVW+
Sbjct: 623 LVGFGLYELGFPGVAKCVEIGLPELILLVFVSQYVPHVLHSGKNIFDRFSVLFTIAIVWI 802
Query: 241 YAIILTGCGAYKNAEHETQDTCRTDRAGLIHGASWISPPIPFRWGAPTFDAGEAFAMMAA 300
YA++LT GAY + +TQ +CRTDRAGLI A WI P PF+WGAPTFDAGEAFAMM A
Sbjct: 803 YAVLLTVGGAYNGSPPKTQTSCRTDRAGLIDAAPWIRVPYPFQWGAPTFDAGEAFAMMMA 982
Query: 301 SFVAQIESTGGFIAVARFASATPVPPSVLSRGIGWQGVGILLSGIFGTGNGSSVSIENAG 360
SFVA +ES+G FIAV RFASATP+PPS+LSRGIGWQGVGILLSG+FGT +GSSVS+ENAG
Sbjct: 983 SFVALVESSGAFIAVYRFASATPLPPSILSRGIGWQGVGILLSGLFGTISGSSVSVENAG 1162
Query: 361 LLALTRVGSRRVVQISAGFMIFFSILGKFGAVFASIPMPIVAALYCLLFSQVGSAGLSFL 420
LLALTRVGSRRVVQISAGFMIFFSILGKFGAVFASIP I+AALYCL F+ VG+ GLSFL
Sbjct: 1163LLALTRVGSRRVVQISAGFMIFFSILGKFGAVFASIPPAIIAALYCLFFAYVGAGGLSFL 1342
Query: 421 QFCNLNSFRTKFIIGFSIFMGFSVPQYFKEYTAIKQYGPVHTNARWFNDMINVPFSSGAF 480
QFCNLNSFRTKFI+GFSIF+G SVPQYF EYTAI YGPVHT RWFNDM+NVPF S AF
Sbjct: 1343QFCNLNSFRTKFILGFSIFLGLSVPQYFNEYTAINGYGPVHTGGRWFNDMVNVPFQSKAF 1522
Query: 481 VAGILALFFDVTLHKSDNQTRKDRGMHWWDRFSSFKTDTRSEEFYSLPFNLNKFFPSV 538
VAG++A F D TLHK D+ RKDRG HWWD++ SFK DTRSEEFYSLPFNLNK+FPSV
Sbjct: 1523VAGVVAYFLDNTLHKRDSSIRKDRGKHWWDKYKSFKGDTRSEEFYSLPFNLNKYFPSV 1696
>TC89841 similar to PIR|T02307|T02307 probable membrane transporter
At2g34190 [imported] - Arabidopsis thaliana, partial
(63%)
Length = 1165
Score = 397 bits (1020), Expect = e-111
Identities = 192/324 (59%), Positives = 236/324 (72%)
Frame = +3
Query: 213 QFMPHMMKGGRHIFARFAVIFSVIIVWVYAIILTGCGAYKNAEHETQDTCRTDRAGLIHG 272
Q++ + K I RFA++ + ++W YA +LT GAYK+ TQ +CRTDRA LI
Sbjct: 27 QYLKNFQKRQVPILERFALLITTTVIWAYAHLLTASGAYKHRPDVTQHSCRTDRANLISS 206
Query: 273 ASWISPPIPFRWGAPTFDAGEAFAMMAASFVAQIESTGGFIAVARFASATPVPPSVLSRG 332
A WI P P WGAPTFDAG +F MMAA V+ +ESTG F A +R ASATP P VLSRG
Sbjct: 207 APWIKIPYPLEWGAPTFDAGHSFGMMAAVLVSLVESTGAFKAASRLASATPPPAHVLSRG 386
Query: 333 IGWQGVGILLSGIFGTGNGSSVSIENAGLLALTRVGSRRVVQISAGFMIFFSILGKFGAV 392
IGWQG+GILL+G+FGT GS+VS+EN GLL RVGSRRV+Q+SAGFMIFF++LGKFGA+
Sbjct: 387 IGWQGIGILLNGLFGTLTGSTVSVENVGLLGSNRVGSRRVIQVSAGFMIFFAMLGKFGAL 566
Query: 393 FASIPMPIVAALYCLLFSQVGSAGLSFLQFCNLNSFRTKFIIGFSIFMGFSVPQYFKEYT 452
FASIP PI AA+YC+LF V S GLSFLQF N+NS R FI G ++F+G S+P+YF+EYT
Sbjct: 567 FASIPFPIFAAIYCVLFGLVASVGLSFLQFTNMNSMRNLFITGVALFLGLSIPEYFREYT 746
Query: 453 AIKQYGPVHTNARWFNDMINVPFSSGAFVAGILALFFDVTLHKSDNQTRKDRGMHWWDRF 512
+GP HT A WFND +N F S VA I+A+F D TL D + KDRGM WW +F
Sbjct: 747 IRALHGPAHTKAGWFNDFLNTIFYSSPTVALIIAVFLDNTLDYKD--SAKDRGMPWWAKF 920
Query: 513 SSFKTDTRSEEFYSLPFNLNKFFP 536
+FK D+R+EEFYSLPFNLN+FFP
Sbjct: 921 RTFKADSRNEEFYSLPFNLNRFFP 992
>TC93725 similar to GP|10177467|dbj|BAB10858. permease 1 {Arabidopsis
thaliana}, partial (48%)
Length = 985
Score = 218 bits (554), Expect = 6e-57
Identities = 108/139 (77%), Positives = 116/139 (82%)
Frame = +3
Query: 7 GGGATPPPKQDEFQPHPVKDQLPNVSYCITSPPPWPEAIMLGFQHYLVMLGTTVLIPTAL 66
GGGA P PK DE PHPVKDQLPNVS+CITSPPPWPEAI+LGFQHYLVMLGTTVLIP++L
Sbjct: 195 GGGAAPQPKLDELLPHPVKDQLPNVSFCITSPPPWPEAILLGFQHYLVMLGTTVLIPSSL 374
Query: 67 VSQMGGGNEEKAMLIQNHLFVAGINTLIQTLFGTRLPAVIGGSFTFVPTTISIILASRYD 126
V QMGGGNEEKA +IQ LFVAGINT QT FGTRLPAVIGGS+TFVPTTISIILA RY
Sbjct: 375 VPQMGGGNEEKAKVIQTLLFVAGINTFFQTTFGTRLPAVIGGSYTFVPTTISIILAGRY- 551
Query: 127 DDIMHPREKFKRIMRGTQG 145
DI++P EKF G G
Sbjct: 552 SDIVNPHEKF*EDNEGNTG 608
Score = 209 bits (533), Expect = 2e-54
Identities = 102/135 (75%), Positives = 121/135 (89%)
Frame = +1
Query: 136 FKRIMRGTQGALIVASSLQIIVGFSGLWCHVVRFISPLSAVPLVALTGFGLYELGFPMLA 195
F++IMRGTQGALIVAS+LQI++GFSGLW +VVRF+SPLSAVPLVAL+GFGLYE GFP+LA
Sbjct: 580 FEKIMRGTQGALIVASTLQIVLGFSGLWRNVVRFLSPLSAVPLVALSGFGLYEFGFPVLA 759
Query: 196 KCIEIGLPEIVILVFLSQFMPHMMKGGRHIFARFAVIFSVIIVWVYAIILTGCGAYKNAE 255
KC+EIGLPEI+ILV SQ++PHMMKG + IF RFAVIFSV IVW+YA +LT GAYKN+
Sbjct: 760 KCVEIGLPEIIILVVFSQYIPHMMKGEKPIFDRFAVIFSVAIVWLYAYLLTVGGAYKNSA 939
Query: 256 HETQDTCRTDRAGLI 270
+TQ TCRTDRAG+I
Sbjct: 940 PKTQITCRTDRAGII 984
>BG452976 weakly similar to GP|15983805|gb At1g49960/F2J10_14 {Arabidopsis
thaliana}, partial (33%)
Length = 610
Score = 212 bits (540), Expect = 3e-55
Identities = 114/209 (54%), Positives = 137/209 (65%)
Frame = +2
Query: 1 MAGGGGGGGATPPPKQDEFQPHPVKDQLPNVSYCITSPPPWPEAIMLGFQHYLVMLGTTV 60
MA G G K D F + V C+ S P WPE I LGFQHYLVMLGT V
Sbjct: 8 MAAGESG-------KVDHFSTSSCQRTTLLVDCCVRSSPSWPEGIFLGFQHYLVMLGTIV 166
Query: 61 LIPTALVSQMGGGNEEKAMLIQNHLFVAGINTLIQTLFGTRLPAVIGGSFTFVPTTISII 120
++ T LV MGGGN EKA +IQ LFVAGINTL+QT GTRLP VIG SF F+ IS+
Sbjct: 167 IVSTILVPLMGGGNVEKAEMIQTLLFVAGINTLLQTWLGTRLPVVIGASFAFIIPAISVA 346
Query: 121 LASRYDDDIMHPREKFKRIMRGTQGALIVASSLQIIVGFSGLWCHVVRFISPLSAVPLVA 180
+SR ++PR++FK+ MR QGAL++AS +Q ++GF G W RF PLSAVPLV
Sbjct: 347 FSSRM-SVFLNPRQRFKQSMRAIQGALLIASFVQGMIGFFGFWGIFGRFFXPLSAVPLVT 523
Query: 181 LTGFGLYELGFPMLAKCIEIGLPEIVILV 209
LTG GL+ +GFP LA C+EIGLP +VILV
Sbjct: 524 LTGLGLFVIGFPRLAXCVEIGLPALVILV 610
>BM815623 similar to GP|14334908|gb putative membrane transporter protein
{Arabidopsis thaliana}, partial (29%)
Length = 461
Score = 200 bits (508), Expect = 1e-51
Identities = 97/150 (64%), Positives = 117/150 (77%)
Frame = +1
Query: 344 GIFGTGNGSSVSIENAGLLALTRVGSRRVVQISAGFMIFFSILGKFGAVFASIPMPIVAA 403
G+FGT G SVS+EN GLL TRVGSRRV+QISAGFMIFFS+LGKFGA FASIP PI AA
Sbjct: 1 GLFGTLTGPSVSVENVGLLGSTRVGSRRVIQISAGFMIFFSMLGKFGAFFASIPFPIFAA 180
Query: 404 LYCLLFSQVGSAGLSFLQFCNLNSFRTKFIIGFSIFMGFSVPQYFKEYTAIKQYGPVHTN 463
+YC+LF V S GLSFLQF N+NS R+ FI G S+F+G S+P+YF+E+T+ +GP HT
Sbjct: 181 MYCVLFGLVASVGLSFLQFTNMNSMRSLFITGVSLFLGLSIPEYFREFTSKALHGPAHTK 360
Query: 464 ARWFNDMINVPFSSGAFVAGILALFFDVTL 493
ARWFND +N F S + VA I+A+F D TL
Sbjct: 361 ARWFNDFLNTIFFSSSTVAFIVAVFLDNTL 450
>BQ149045 similar to PIR|T02307|T02 probable membrane transporter At2g34190
[imported] - Arabidopsis thaliana, partial (38%)
Length = 650
Score = 177 bits (448), Expect = 1e-44
Identities = 87/191 (45%), Positives = 116/191 (60%)
Frame = +2
Query: 129 IMHPREKFKRIMRGTQGALIVASSLQIIVGFSGLWCHVVRFISPLSAVPLVALTGFGLYE 188
I P +F + MR QGA+I ASS+QII+GFS LW R SPL VP++AL+GFGL+
Sbjct: 44 IEDPHLRFVKTMRAVQGAMIAASSIQIILGFSQLWAICSRXFSPLGMVPVIALSGFGLFN 223
Query: 189 LGFPMLAKCIEIGLPEIVILVFLSQFMPHMMKGGRHIFARFAVIFSVIIVWVYAIILTGC 248
GFP++ C+E G+P +++ V SQ++ + I RFA++ S I+W YA +LT
Sbjct: 224 RGFPVVGHCVETGIPMLILXVAFSQYLKNFNARQLPILERFALLISTTIIWAYAHLLTTS 403
Query: 249 GAYKNAEHETQDTCRTDRAGLIHGASWISPPIPFRWGAPTFDAGEAFAMMAASFVAQIES 308
G YK TQ CRTD++ LI W+ P PF G PTFD G AF MM A V+ IES
Sbjct: 404 GTYKLRSELTQYNCRTDKSNLISSXPWMKIPYPFEXGFPTFDXGHAFGMMXAVLVSLIES 583
Query: 309 TGGFIAVARFA 319
TG + A + A
Sbjct: 584 TGAYTAASXLA 616
>BQ124592 similar to GP|20466159|gb putative membrane transporter
{Arabidopsis thaliana}, partial (32%)
Length = 690
Score = 154 bits (388), Expect = 1e-37
Identities = 74/190 (38%), Positives = 112/190 (58%)
Frame = +2
Query: 262 CRTDRAGLIHGASWISPPIPFRWGAPTFDAGEAFAMMAASFVAQIESTGGFIAVARFASA 321
CR D + I + W P P +WG P F A M S ++ ++S G + A + ++
Sbjct: 11 CRVDTSHAIKSSPWFKFPYPLQWGTPVFHWKMALVMCVVSLISSVDSVGSYHASSLLVAS 190
Query: 322 TPVPPSVLSRGIGWQGVGILLSGIFGTGNGSSVSIENAGLLALTRVGSRRVVQISAGFMI 381
P P VLSRGIG +G+ +L+G++GTG GS+ EN +A+T++GSRR VQ+ A +I
Sbjct: 191 RPPTPGVLSRGIGLEGLSSVLAGLWGTGTGSTTLTENVHTIAVTKMGSRRAVQLGACILI 370
Query: 382 FFSILGKFGAVFASIPMPIVAALYCLLFSQVGSAGLSFLQFCNLNSFRTKFIIGFSIFMG 441
S++GK G ASIP +VA L C +++ + + GLS L++ S R IIG S+F
Sbjct: 371 VLSLVGKVGGFIASIPGVMVAGLLCFMWAMLTALGLSNLRYSEAGSSRNIIIIGLSLFFS 550
Query: 442 FSVPQYFKEY 451
S+P YF++Y
Sbjct: 551 LSIPAYFQQY 580
>TC89886 weakly similar to GP|14587294|dbj|BAB61205. putative permease 1
{Oryza sativa (japonica cultivar-group)}, partial (25%)
Length = 638
Score = 151 bits (381), Expect = 7e-37
Identities = 77/158 (48%), Positives = 98/158 (61%)
Frame = +1
Query: 380 MIFFSILGKFGAVFASIPMPIVAALYCLLFSQVGSAGLSFLQFCNLNSFRTKFIIGFSIF 439
MI SI GKFGA FASIP+PI AA+Y +LF V + +SF+QF N NS R ++ G ++F
Sbjct: 10 MILCSIFGKFGAFFASIPLPIFAAIY*VLFGIVAATRISFIQFANNNSIRNIYVFGLTLF 189
Query: 440 MGFSVPQYFKEYTAIKQYGPVHTNARWFNDMINVPFSSGAFVAGILALFFDVTLHKSDNQ 499
+G S+PQYF TA +GPV TN WFND++N FSS VA I+ D TL Q
Sbjct: 190 LGISIPQYFVMNTAPDGHGPVRTNGGWFNDILNTIFSSPPTVAIIVGTVLDNTLEA--KQ 363
Query: 500 TRKDRGMHWWDRFSSFKTDTRSEEFYSLPFNLNKFFPS 537
T DRG+ WW F K D R++EFY P L ++ PS
Sbjct: 364 TAVDRGLPWWVPFQKRKGDVRNDEFYRFPLRLTEYIPS 477
>BF637188 similar to GP|14334908|gb putative membrane transporter protein
{Arabidopsis thaliana}, partial (19%)
Length = 448
Score = 118 bits (295), Expect = 7e-27
Identities = 54/92 (58%), Positives = 68/92 (73%)
Frame = +2
Query: 22 HPVKDQLPNVSYCITSPPPWPEAIMLGFQHYLVMLGTTVLIPTALVSQMGGGNEEKAMLI 81
H DQL + YCI S P W E I+LGFQHY++ LGT V+IP+ LV MGG +++K ++
Sbjct: 161 HSPMDQLQGLEYCIDSNPSWVETILLGFQHYILALGTAVMIPSFLVPSMGGNDDDKVRVV 340
Query: 82 QNHLFVAGINTLIQTLFGTRLPAVIGGSFTFV 113
Q LFV GINTL+QTLFGTRLP VIGGS+ F+
Sbjct: 341 QTLLFVEGINTLLQTLFGTRLPTVIGGSYAFM 436
>CA990413 similar to PIR|T05632|T05 hypothetical protein F20D10.170 -
Arabidopsis thaliana, partial (22%)
Length = 731
Score = 105 bits (261), Expect = 6e-23
Identities = 60/180 (33%), Positives = 93/180 (51%), Gaps = 14/180 (7%)
Frame = +1
Query: 366 RVGSRRVVQISAGFMIFFSILGKFGAVFASIPMPIVAALYCLLFSQVGSAGLSFLQFCNL 425
+V SRRV+++ A F+I FS +GK GA+ ASIP + AA+ C +++ + GLS LQ+
Sbjct: 1 KVASRRVLELGAVFLILFSFVGKVGALLASIPQALAAAILCFMWALTVALGLSTLQYGQS 180
Query: 426 NSFRTKFIIGFSIFMGFSVPQYFKEYT--------------AIKQYGPVHTNARWFNDMI 471
SFR I+G ++F+G S+P YF++Y A GP H+ + + I
Sbjct: 181 PSFRNMTIVGVALFLGMSIPSYFQQYQPESSLILPSYLVPYAAASSGPFHSGLKQLDFAI 360
Query: 472 NVPFSSGAFVAGILALFFDVTLHKSDNQTRKDRGMHWWDRFSSFKTDTRSEEFYSLPFNL 531
N S V ++A D T+ S +++RG++ W R D + YSLP L
Sbjct: 361 NALMSMNMVVTLLVAFLLDNTVPGS----KQERGVYTWSRAEDIAADASLQSEYSLPKKL 528
>AW697093 similar to GP|20466159|gb putative membrane transporter
{Arabidopsis thaliana}, partial (18%)
Length = 603
Score = 56.2 bits (134), Expect(2) = 5e-17
Identities = 24/55 (43%), Positives = 37/55 (66%)
Frame = +1
Query: 48 GFQHYLVMLGTTVLIPTALVSQMGGGNEEKAMLIQNHLFVAGINTLIQTLFGTRL 102
G QHY+ MLG+ +LIP +V MGG +EE + ++ LFV+G+ TL+ FG+ +
Sbjct: 235 GIQHYVSMLGSLILIPLVIVPAMGGSHEETSNVVSTVLFVSGLTTLLHISFGSEI 399
Score = 49.7 bits (117), Expect(2) = 5e-17
Identities = 24/72 (33%), Positives = 44/72 (60%)
Frame = +2
Query: 97 LFGTRLPAVIGGSFTFVPTTISIILASRYDDDIMHPREKFKRIMRGTQGALIVASSLQII 156
+ G RLP + G SF ++ ++II + ++ +KFK IMR QGA+I+ S+ Q +
Sbjct: 383 VLGQRLPLIQGPSFVYLAPALAIINSPELQG--LNGNDKFKHIMRELQGAIIIGSAFQAL 556
Query: 157 VGFSGLWCHVVR 168
+G++GL +++
Sbjct: 557 LGYTGLMSLLIK 592
>AW689217 similar to GP|15144504|gb| putative permease {Lycopersicon
esculentum}, partial (16%)
Length = 609
Score = 64.7 bits (156), Expect = 9e-11
Identities = 32/89 (35%), Positives = 50/89 (55%)
Frame = +2
Query: 30 NVSYCITSPPPWPEAIMLGFQHYLVMLGTTVLIPTALVSQMGGGNEEKAMLIQNHLFVAG 89
++ + +T P + QHYL ++G+ VLIP +V MGG + + A +I LF++G
Sbjct: 266 DMKFGLTENPGLVPLVYYSLQHYLSLIGSLVLIPLVMVPTMGGTDNDTANVISTMLFLSG 445
Query: 90 INTLIQTLFGTRLPAVIGGSFTFVPTTIS 118
I T++ + FGTRLP V G T+ S
Sbjct: 446 ITTILHSYFGTRLPLVQGSLICVFSTSTS 532
>AW695592 weakly similar to PIR|T00984|T009 probable membrane transporter
At2g26510 [imported] - Arabidopsis thaliana, partial
(10%)
Length = 173
Score = 60.8 bits (146), Expect = 1e-09
Identities = 26/55 (47%), Positives = 37/55 (67%)
Frame = +1
Query: 228 RFAVIFSVIIVWVYAIILTGCGAYKNAEHETQDTCRTDRAGLIHGASWISPPIPF 282
RFA++ + I+W +A ILT GAY ++ +TQ +CRTDR+ L+ A WI P PF
Sbjct: 7 RFALLICIAIIWAFAAILTVAGAYNTSKEKTQTSCRTDRSYLLTRAPWIYVPYPF 171
>CB892373 PIR|D64842|D64 probable transport protein ycdG - Escherichia coli,
partial (21%)
Length = 526
Score = 38.9 bits (89), Expect = 0.005
Identities = 20/68 (29%), Positives = 35/68 (51%)
Frame = +2
Query: 349 GNGSSVSIENAGLLALTRVGSRRVVQISAGFMIFFSILGKFGAVFASIPMPIVAALYCLL 408
G+G + EN G++A+T+V S V +A + KFGA+ +IP ++ ++
Sbjct: 8 GSGVTTYAENIGVMAVTKVYSTLVFVAAAVIAMLLGFSPKFGALIHTIPAAVIGGASIVV 187
Query: 409 FSQVGSAG 416
F + AG
Sbjct: 188 FGLIAVAG 211
>AW586210
Length = 625
Score = 32.7 bits (73), Expect = 0.36
Identities = 14/29 (48%), Positives = 17/29 (58%)
Frame = +2
Query: 510 DRFSSFKTDTRSEEFYSLPFNLNKFFPSV 538
D F SF T +SL FNLNK+FP +
Sbjct: 179 DSFKSFLTYNHPSRLFSLSFNLNKWFPHI 265
>BQ135858 similar to GP|23491723|db formin homology protein A {Dictyostelium
discoideum}, partial (4%)
Length = 1033
Score = 32.3 bits (72), Expect = 0.47
Identities = 18/47 (38%), Positives = 22/47 (46%), Gaps = 5/47 (10%)
Frame = -2
Query: 4 GGGGGGATPPPKQ-----DEFQPHPVKDQLPNVSYCITSPPPWPEAI 45
GGGGGG PPP D P P K ++P PPP P+ +
Sbjct: 513 GGGGGGVPPPPPPPGGGGDPPPPPPPKKKIP-------PPPPPPDGV 394
Score = 29.3 bits (64), Expect = 4.0
Identities = 18/45 (40%), Positives = 20/45 (44%), Gaps = 4/45 (8%)
Frame = -3
Query: 2 AGGGGGGGATPPPKQ----DEFQPHPVKDQLPNVSYCITSPPPWP 42
AGGGGGG PPP + P P K + P PPP P
Sbjct: 515 AGGGGGGSPPPPPPRGGGGTPPPPPPPKKKFP--------PPPPP 405
Score = 29.3 bits (64), Expect = 4.0
Identities = 10/13 (76%), Positives = 11/13 (83%)
Frame = +1
Query: 3 GGGGGGGATPPPK 15
GGGGGGG PPP+
Sbjct: 436 GGGGGGGVPPPPR 474
Score = 28.5 bits (62), Expect = 6.8
Identities = 10/12 (83%), Positives = 10/12 (83%)
Frame = +2
Query: 3 GGGGGGGATPPP 14
GGGGGGG PPP
Sbjct: 473 GGGGGGGTPPPP 508
>AL384532
Length = 364
Score = 31.6 bits (70), Expect = 0.80
Identities = 14/40 (35%), Positives = 24/40 (60%)
Frame = +2
Query: 498 NQTRKDRGMHWWDRFSSFKTDTRSEEFYSLPFNLNKFFPS 537
++T KDR H++ F FKT++ + + +L FN+N S
Sbjct: 245 SRTSKDRLFHYFINFPYFKTNSNIKNYLNLFFNINHIINS 364
>BQ144539 GP|13173388|g lysine/glutamic acid-rich protein {Candida albicans},
partial (1%)
Length = 476
Score = 30.4 bits (67), Expect = 1.8
Identities = 21/63 (33%), Positives = 34/63 (53%)
Frame = -1
Query: 383 FSILGKFGAVFASIPMPIVAALYCLLFSQVGSAGLSFLQFCNLNSFRTKFIIGFSIFMGF 442
F IL F +F I+A+ + LLF ++ ++ SFL + +S FI GF +F+ F
Sbjct: 236 FRILASFFFIFF---F*ILASFFSLLFFRISASSFSFL---SGSSSSFLFISGFLLFLPF 75
Query: 443 SVP 445
+P
Sbjct: 74 RLP 66
>TC90405 similar to GP|18086395|gb|AAL57656.1 AT5g13210/T31B5_30
{Arabidopsis thaliana}, partial (32%)
Length = 897
Score = 29.6 bits (65), Expect = 3.1
Identities = 13/33 (39%), Positives = 20/33 (60%)
Frame = -1
Query: 10 ATPPPKQDEFQPHPVKDQLPNVSYCITSPPPWP 42
++ PP Q HP+ DQ+P+ + C +SP P P
Sbjct: 429 SSDPP*DSHLQRHPICDQIPSGNQC-SSPNPSP 334
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.327 0.142 0.443
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 17,187,527
Number of Sequences: 36976
Number of extensions: 249637
Number of successful extensions: 3555
Number of sequences better than 10.0: 55
Number of HSP's better than 10.0 without gapping: 2217
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 3422
length of query: 538
length of database: 9,014,727
effective HSP length: 101
effective length of query: 437
effective length of database: 5,280,151
effective search space: 2307425987
effective search space used: 2307425987
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 15 ( 7.1 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.7 bits)
S2: 61 (28.1 bits)
Medicago: description of AC126785.11


