

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC121237.19 + phase: 0 /pseudo
(754 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
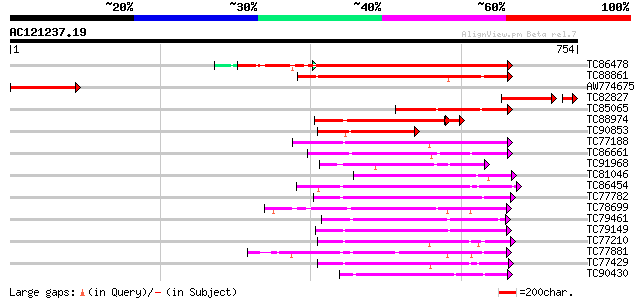
Score E
Sequences producing significant alignments: (bits) Value
TC86478 similar to GP|3688193|emb|CAA08995.1 MAP3K alpha 1 prote... 429 e-120
TC88861 weakly similar to GP|17027283|gb|AAL34137.1 putative pro... 299 3e-81
AW774675 weakly similar to PIR|A96662|A966 hypothetical protein ... 196 2e-50
TC82827 similar to PIR|A96662|A96662 hypothetical protein F24D7.... 147 3e-41
TC85065 similar to GP|1255448|dbj|BAA09057.1 mitogen-activated p... 164 1e-40
TC88974 similar to GP|3819699|emb|CAA08758.1 BnMAP4K alpha2 {Bra... 136 7e-35
TC90853 similar to GP|2342427|dbj|BAA21857.1 NPK1-related protei... 141 8e-34
TC77188 similar to PIR|T04862|T04862 probable serine/threonine-s... 140 2e-33
TC86661 similar to GP|9798599|dbj|BAB11737.1 serine/threonine pr... 135 8e-32
TC91968 similar to PIR|T02550|T02550 NPK1-related protein kinase... 134 1e-31
TC81046 similar to GP|17529280|gb|AAL38867.1 putative protein ki... 134 2e-31
TC86454 homologue to GP|4567091|gb|AAD23582.1| SNF-1-like serine... 133 2e-31
TC77782 similar to GP|13448037|gb|AAK26845.1 SOS2-like protein k... 127 1e-29
TC78699 weakly similar to PIR|G96761|G96761 probable MAP kinase ... 127 2e-29
TC79461 similar to GP|9758254|dbj|BAB08753.1 contains similarity... 127 2e-29
TC79149 similar to PIR|T46059|T46059 MAP kinase - Arabidopsis th... 127 2e-29
TC77210 similar to GP|21954717|gb|AAM83095.1 SOS2-like protein k... 126 3e-29
TC77881 homologue to GP|15528439|emb|CAC69137. MEK map kinase ki... 125 8e-29
TC77429 similar to GP|15215664|gb|AAK91377.1 AT5g25110/T11H3_120... 124 1e-28
TC90430 similar to GP|18766642|gb|AAL79042.1 NIMA-related protei... 123 3e-28
>TC86478 similar to GP|3688193|emb|CAA08995.1 MAP3K alpha 1 protein kinase
{Brassica napus}, partial (52%)
Length = 2266
Score = 429 bits (1103), Expect = e-120
Identities = 217/365 (59%), Positives = 270/365 (73%)
Frame = +2
Query: 304 SGPLFWQPSRGSPEYSPIPSPRMTSPGPSSRIQSGAVTPIHPRAGGTPTESQTGRADDGK 363
+ PL +RG + P T GP+S +P+HPR G +S TG+ DDG+
Sbjct: 701 TSPLHPNATRGPTSPTSPLHPNATR-GPTS-----PTSPLHPRLQGLNLDSPTGKQDDGR 862
Query: 364 QQSHRLPLPPLTVTNTSPFSHSNSAATSPSMPRSPARADSPMSSGSRWKKGKLLGRGTFG 423
Q H LPLPP SP S S++ + + RSP SP + S+WKKGKLLGRGTFG
Sbjct: 863 SQCHPLPLPP-----GSPTSPSSALSNT----RSPFENSSP--NLSKWKKGKLLGRGTFG 1009
Query: 424 HVYIGFNSQSGEMCAMKEVTLFSDDAKSLESAKQLMQEVHLLSRLRHPNIVQYYGSETVD 483
HVY+GFNS++G+MCA+KEV + DD S E KQL QE+ LL++L HPNIVQY GSE +
Sbjct: 1010 HVYLGFNSENGQMCAIKEVRVGCDDQNSKECLKQLHQEIDLLNQLSHPNIVQYLGSELGE 1189
Query: 484 DKLYIYLEYVSGGSIHKLLQEYGQFGELAIRSYTQQILSGLAYLHAKNTLHRDIKGANIL 543
+ L +YLEYVSGGSIHKLLQEYG F E I++YT+QI+SGLAYLH +NT+HRDIKGANIL
Sbjct: 1190 ESLSVYLEYVSGGSIHKLLQEYGPFKEPVIQNYTRQIVSGLAYLHGRNTVHRDIKGANIL 1369
Query: 544 VDPNGRVKVADFGMAKHITGQYCPLSFKGSPYWMAPEVIKNSKECSLGVDIWSLGCTVLE 603
VDPNG +K+ADFGMAKHIT LSFKGSPYWMAPEV+ N+ SL VDIWSLGCT++E
Sbjct: 1370 VDPNGEIKLADFGMAKHITSAASMLSFKGSPYWMAPEVVMNTNGYSLPVDIWSLGCTLIE 1549
Query: 604 MATTKPPWSQYEGVAAMFKIGNSKELPTIPDHLSNEGKDFVRKCLQRNPRDRPSASELLD 663
MA +KPPWSQYEGVAA+FKIGNSK++P IP+HLSN+ K+F+ CLQR+P RP+A +LL+
Sbjct: 1550 MAASKPPWSQYEGVAAIFKIGNSKDMPIIPEHLSNDAKNFIMLCLQRDPSARPTAQKLLE 1729
Query: 664 HPFVK 668
HPF++
Sbjct: 1730 HPFIR 1744
Score = 42.0 bits (97), Expect = 9e-04
Identities = 46/145 (31%), Positives = 57/145 (38%), Gaps = 8/145 (5%)
Frame = +2
Query: 273 PYPEINFVGSGHCSSPGSGHNSGHNSMGGDMSGPLFWQPSRGSPEYSPIPSPRMTSPGPS 332
P P + G S GS +S S P F +RG E + R SPGP+
Sbjct: 374 PLPVNSDQALGSFSVSGSSVSSS-TSFDDHPISPHFISNNRGQDEVKF--NVRSRSPGPA 544
Query: 333 SRIQSGAVTPIHPRAGGTPTESQTGRADDGKQQSHRLPLPPL----TVTNTSPFS--HSN 386
SR + +P+HP A PT S T + P PL T TSP S H N
Sbjct: 545 SRGPTSPTSPLHPNASRGPT-SPTSPLHPAASRGPTSPTSPLHPNATRGPTSPTSPLHPN 721
Query: 387 S--AATSPSMPRSPARADSPMSSGS 409
+ TSP+ P P P S S
Sbjct: 722 ATRGPTSPTSPLHPNATRGPTSPTS 796
>TC88861 weakly similar to GP|17027283|gb|AAL34137.1 putative protein kinase
{Oryza sativa (japonica cultivar-group)}, partial (34%)
Length = 1847
Score = 299 bits (765), Expect = 3e-81
Identities = 151/293 (51%), Positives = 204/293 (69%), Gaps = 7/293 (2%)
Frame = +1
Query: 383 SHSNSAATSPSMPRSPARADSPMSSGSRWKKGKLLGRGTFGHVYIGFNSQSGEMCAMKEV 442
SH+N + S+ + + + G W+KGKL+GRG+FG VY N ++G CA+KEV
Sbjct: 943 SHNNHLHINSSVTMNHSTENIHSMKG-HWQKGKLIGRGSFGSVYHATNLETGASCALKEV 1119
Query: 443 TLFSDDAKSLESAKQLMQEVHLLSRLRHPNIVQYYGSETVDDKLYIYLEYVSGGSIHKLL 502
L DD KS + KQL QE+ +L +L HPNIV+YYGSE V D+L IY+EYV GS+ K +
Sbjct: 1120 DLVPDDPKSTDCIKQLDQEIRILGQLHHPNIVEYYGSEVVGDRLCIYMEYVHPGSLQKFM 1299
Query: 503 QEY-GQFGELAIRSYTQQILSGLAYLHAKNTLHRDIKGANILVDPNGRVKVADFGMAKHI 561
Q++ G E +R++T+ ILSGLAYLH+ T+HRDIKGAN+LVD +G VK+ADFG++K +
Sbjct: 1300 QDHCGVMTESVVRNFTRHILSGLAYLHSTKTIHRDIKGANLLVDASGIVKLADFGVSKIL 1479
Query: 562 TGQYCPLSFKGSPYWMAPEVI------KNSKECSLGVDIWSLGCTVLEMATTKPPWSQYE 615
T + LS KGSPYWMAPE++ + + ++ VDIWSLGCT++EM T KPPWS++
Sbjct: 1480 TEKSYELSLKGSPYWMAPELMMAAMKNETNPTVAMAVDIWSLGCTIIEMLTGKPPWSEFP 1659
Query: 616 GVAAMFKIGNSKELPTIPDHLSNEGKDFVRKCLQRNPRDRPSASELLDHPFVK 668
G AMFK+ + P IP LS EG+DF+ +C QRNP DRPSA+ LL HPFV+
Sbjct: 1660 GHQAMFKVLHRS--PDIPKTLSPEGQDFLEQCFQRNPADRPSAAVLLTHPFVQ 1812
>AW774675 weakly similar to PIR|A96662|A966 hypothetical protein F24D7.11
[imported] - Arabidopsis thaliana, partial (9%)
Length = 632
Score = 196 bits (499), Expect = 2e-50
Identities = 94/94 (100%), Positives = 94/94 (100%)
Frame = +2
Query: 1 MPTWWGKSSSKETKKKAGKESIIDTLHRKFKFPSEGKRSTISGGSRRRSNDTISEKGDRS 60
MPTWWGKSSSKETKKKAGKESIIDTLHRKFKFPSEGKRSTISGGSRRRSNDTISEKGDRS
Sbjct: 350 MPTWWGKSSSKETKKKAGKESIIDTLHRKFKFPSEGKRSTISGGSRRRSNDTISEKGDRS 529
Query: 61 PSESRSPSPSKVARCQSFAERPHAQPLPLPDLHP 94
PSESRSPSPSKVARCQSFAERPHAQPLPLPDLHP
Sbjct: 530 PSESRSPSPSKVARCQSFAERPHAQPLPLPDLHP 631
>TC82827 similar to PIR|A96662|A96662 hypothetical protein F24D7.11
[imported] - Arabidopsis thaliana, partial (3%)
Length = 1102
Score = 147 bits (370), Expect(2) = 3e-41
Identities = 72/73 (98%), Positives = 72/73 (98%)
Frame = +3
Query: 655 RPSASELLDHPFVKGAAPLERPIMVPEASDPITGITHGTKALGIGQGRNLSALDSDKLLV 714
RPS SELLDHPFVKGAAPLERPIMVPEASDPITGITHGTKALGIGQGRNLSALDSDKLLV
Sbjct: 3 RPSVSELLDHPFVKGAAPLERPIMVPEASDPITGITHGTKALGIGQGRNLSALDSDKLLV 182
Query: 715 HSSRVLKNNPHES 727
HSSRVLKNNPHES
Sbjct: 183 HSSRVLKNNPHES 221
Score = 40.8 bits (94), Expect(2) = 3e-41
Identities = 19/19 (100%), Positives = 19/19 (100%)
Frame = +1
Query: 736 VKSIFKGIYLAQSLPLEAH 754
VKSIFKGIYLAQSLPLEAH
Sbjct: 220 VKSIFKGIYLAQSLPLEAH 276
>TC85065 similar to GP|1255448|dbj|BAA09057.1 mitogen-activated protein
kinase {Arabidopsis thaliana}, partial (26%)
Length = 789
Score = 164 bits (415), Expect = 1e-40
Identities = 85/157 (54%), Positives = 113/157 (71%), Gaps = 1/157 (0%)
Frame = +2
Query: 513 IRSYTQQILSGLAYLHAKNTLHRDIKGANILVDPNGRVKVADFGMAKHITGQYCPLSFKG 572
+ +YT+QIL GL YLH +N +HRDIK ANILVD NG VKVADFG++ I S +G
Sbjct: 26 VSAYTRQILHGLKYLHDRNIVHRDIKCANILVDANGSVKVADFGLS*AIKLNDVK-SCQG 202
Query: 573 SPYWMAPEVIKNS-KECSLGVDIWSLGCTVLEMATTKPPWSQYEGVAAMFKIGNSKELPT 631
+ +WMAPEV++ K L DIWSLGCTVLEM T + P++ E ++AMF+IG ELP
Sbjct: 203 TAFWMAPEVVRGKVKGYGLPADIWSLGCTVLEMLTGQVPYAPMECISAMFRIGKG-ELPP 379
Query: 632 IPDHLSNEGKDFVRKCLQRNPRDRPSASELLDHPFVK 668
+PD LS + +DF+ +CL+ NP DRP+A++LLDH FV+
Sbjct: 380 VPDTLSRDARDFILQCLKVNPDDRPTAAQLLDHKFVQ 490
>TC88974 similar to GP|3819699|emb|CAA08758.1 BnMAP4K alpha2 {Brassica
napus}, partial (34%)
Length = 851
Score = 136 bits (342), Expect(2) = 7e-35
Identities = 73/182 (40%), Positives = 114/182 (62%), Gaps = 1/182 (0%)
Frame = +3
Query: 406 SSGSRWKKGKLLGRGTFGHVYIGFNSQSGEMCAMKEVTLFSDDAKSLESAKQLMQEVHLL 465
++G+R+ +L+G+G+FG VY GF+ + + A+K + L +S + + +E+ +L
Sbjct: 174 ATGTRFTSLELIGQGSFGDVYKGFDKELNKEVAIKVIDL----EESEDDIDDIQKEISVL 341
Query: 466 SRLRHPNIVQYYGSETVDDKLYIYLEYVSGGSIHKLLQEYGQFGELAIRSYTQQILSGLA 525
S+ R P I +YYGS KL+I +EY++GGS+ LLQ E++I + +L +
Sbjct: 342 SQCRCPYITEYYGSFLNQTKLWIIMEYMAGGSVADLLQSGPPLDEMSIAYILRDLLHAVD 521
Query: 526 YLHAKNTLHRDIKGANILVDPNGRVKVADFGMAKHITGQYC-PLSFKGSPYWMAPEVIKN 584
YLH + +HRDIK ANIL+ NG VKVADFG++ +T +F G+P+WMAPEVI+N
Sbjct: 522 YLHNEGKIHRDIKAANILLSENGDVKVADFGVSAQLTRTISRRKTFVGTPFWMAPEVIQN 701
Query: 585 SK 586
K
Sbjct: 702 LK 707
Score = 30.0 bits (66), Expect(2) = 7e-35
Identities = 13/27 (48%), Positives = 17/27 (62%)
Frame = +1
Query: 579 PEVIKNSKECSLGVDIWSLGCTVLEMA 605
P + + SK + DIWSLG T +EMA
Sbjct: 685 PRLFRTSKGYNEKADIWSLGITAIEMA 765
>TC90853 similar to GP|2342427|dbj|BAA21857.1 NPK1-related protein kinase 3
{Arabidopsis thaliana}, partial (22%)
Length = 746
Score = 141 bits (356), Expect = 8e-34
Identities = 71/141 (50%), Positives = 97/141 (68%), Gaps = 5/141 (3%)
Frame = +3
Query: 410 RWKKGKLLGRGTFGHVYIGFNSQSGEMCAMKEVTL-----FSDDAKSLESAKQLMQEVHL 464
RW+KG+L+G G FG VY+G N SGE+ A+K+V + F ++ K+ + ++L +EV L
Sbjct: 258 RWRKGELIGSGAFGRVYMGMNLDSGELIAVKQVLIEPGIAFKENTKA--NIRELEEEVKL 431
Query: 465 LSRLRHPNIVQYYGSETVDDKLYIYLEYVSGGSIHKLLQEYGQFGELAIRSYTQQILSGL 524
L L+HPNIV+Y G+ +D L I LE+V GGSI LL ++G F E I +YT+Q+L GL
Sbjct: 432 LKNLKHPNIVRYLGTAREEDSLNILLEFVPGGSISSLLGKFGSFPESGITTYTKQLLDGL 611
Query: 525 AYLHAKNTLHRDIKGANILVD 545
YLH +HRDIKGANILVD
Sbjct: 612 EYLHNNRIIHRDIKGANILVD 674
>TC77188 similar to PIR|T04862|T04862 probable serine/threonine-specific
protein kinase (EC 2.7.1.-) F28A21.110 - Arabidopsis
thaliana, partial (79%)
Length = 1881
Score = 140 bits (353), Expect = 2e-33
Identities = 90/295 (30%), Positives = 146/295 (48%), Gaps = 3/295 (1%)
Frame = +1
Query: 377 TNTSPFSHSNSAATSPSMPRSPARADSPMSSGSRWKKGKLLGRGTFGHVYIGFNSQSGEM 436
T ++P S + +P S + D+P R++ GKLLG GTF V++ N ++GE
Sbjct: 88 TLSTPDYPSMAVVAAPKKNNSMNKKDNPNLLLGRFELGKLLGHGTFAKVHLAKNLKTGES 267
Query: 437 CAMKEVTLFSDDAKSLESAKQLMQEVHLLSRLRHPNIVQYYGSETVDDKLYIYLEYVSGG 496
A+K ++ D + +E+ +L R+RHPNIVQ + K+Y +EYV GG
Sbjct: 268 VAIKIIS--KDKILKSGLVSHIKREISILRRVRHPNIVQLFEVMATKTKIYFVMEYVRGG 441
Query: 497 SIHKLLQEYGQFGELAIRSYTQQILSGLAYLHAKNTLHRDIKGANILVDPNGRVKVADFG 556
+ + + G+ E R Y QQ++ + + HA+ HRD+K N+L+D G +KV+DFG
Sbjct: 442 ELFNKVAK-GRLKEEVARKYFQQLICAVGFCHARGVFHRDLKPENLLLDEKGNLKVSDFG 618
Query: 557 M---AKHITGQYCPLSFKGSPYWMAPEVIKNSKECSLGVDIWSLGCTVLEMATTKPPWSQ 613
+ + I +F G+P ++APEV+ VDIWS G + + P+
Sbjct: 619 LSAVSDEIKQDGLFHTFCGTPAYVAPEVLSRKGYDGAKVDIWSCGVVLFVLMAGYLPFHD 798
Query: 614 YEGVAAMFKIGNSKELPTIPDHLSNEGKDFVRKCLQRNPRDRPSASELLDHPFVK 668
V AM+K E P S E + + L P+ R S E++++ + K
Sbjct: 799 PNNVMAMYKKIYKGEF-RCPRWFSPELVSLLTRLLDIKPQTRISIPEIMENRWFK 960
>TC86661 similar to GP|9798599|dbj|BAB11737.1 serine/threonine protein
kinase {Arabidopsis thaliana}, partial (58%)
Length = 1840
Score = 135 bits (339), Expect = 8e-32
Identities = 84/277 (30%), Positives = 141/277 (50%), Gaps = 5/277 (1%)
Frame = +2
Query: 397 SPARADSPMSSGSRWKKGKLLGRGTFGHVYIGFNSQSGEMCAMKEVTLFSDDAKSLESAK 456
+PA AD+ + +++ GKLLG G F VY N+++G+ A+K + A L A
Sbjct: 263 APAPADATTALFGKYELGKLLGCGAFAKVYHARNTENGQSVAVKVINKKKISATGL--AG 436
Query: 457 QLMQEVHLLSRLRHPNIVQYYGSETVDDKLYIYLEYVSGGSIHKLLQEYGQFGELAIRSY 516
+ +E+ ++S+LRHPNIV+ + K+Y +E+ GG + + G+F E R
Sbjct: 437 HVKREISIMSKLRHPNIVRLHEVLATKTKIYFVMEFAKGGELFAKIANKGRFSEDLSRRL 616
Query: 517 TQQILSGLAYLHAKNTLHRDIKGANILVDPNGRVKVADFGMAK-----HITGQYCPLSFK 571
QQ++S + Y H+ HRD+K N+L+D G +KV+DFG++ I G L
Sbjct: 617 FQQLISAVGYCHSHGVFHRDLKPENLLLDDKGNLKVSDFGLSAVKEQVRIDGMLHTLC-- 790
Query: 572 GSPYWMAPEVIKNSKECSLGVDIWSLGCTVLEMATTKPPWSQYEGVAAMFKIGNSKELPT 631
G+P ++APE++ VDIWS G + + P++ + M++ S E
Sbjct: 791 GTPAYVAPEILAKRGYDGAKVDIWSCGIILFVLVAGYLPFND-PNLMVMYRKIYSGEF-K 964
Query: 632 IPDHLSNEGKDFVRKCLQRNPRDRPSASELLDHPFVK 668
P S + K F+ + + NP R + E+L P+ +
Sbjct: 965 CPRWFSPDLKRFLSRLMDTNPETRITVDEILRDPWFR 1075
>TC91968 similar to PIR|T02550|T02550 NPK1-related protein kinase homolog
T26B15.7 - Arabidopsis thaliana, partial (20%)
Length = 662
Score = 134 bits (337), Expect = 1e-31
Identities = 78/230 (33%), Positives = 127/230 (54%), Gaps = 5/230 (2%)
Frame = +2
Query: 413 KGKLLGRGTFGHVYIGFNSQSGEMCAMKEVTLFSDDAKSLESAKQLMQEVHLLSRLRHPN 472
+G+++GRG+ VY+ SG +F+ + L ++ L +E +L++L+ P
Sbjct: 5 RGRIIGRGSTATVYVADRCHSGG-------EVFAVKSTELHRSELLKKEQQILTKLKCPQ 163
Query: 473 IVQYYGSETVDDK----LYIYLEYVSGGSIHKLLQEYGQFGELAIRSYTQQILSGLAYLH 528
IV Y G E + +++EY G++ ++ E + Y +QIL GL YLH
Sbjct: 164 IVSYQGCEVTFENGVQLFNLFMEYAPKGTLSDAVRNGNGMEEAMVGFYARQILLGLNYLH 343
Query: 529 AKNTLHRDIKGANILVDPNGRVKVADFGMAKHITGQYCPLSFKGSPYWMAPEVIKNSKEC 588
+H D+KG N+LV G K++DFG A+ + + L G+P +MAPEV + +E
Sbjct: 344 LNGIVHCDLKGQNVLVTEQG-AKISDFGCARRVEEE---LVISGTPAFMAPEVAR-GEEQ 508
Query: 589 SLGVDIWSLGCTVLEMATTKPPWSQYEGVAA-MFKIGNSKELPTIPDHLS 637
D+W+LGCT+LEM T K PW + AA +++IG S ++P IPD++S
Sbjct: 509 GYASDVWALGCTLLEMITGKMPWQGFSDPAAVLYRIGFSGDVPEIPDYVS 658
>TC81046 similar to GP|17529280|gb|AAL38867.1 putative protein kinase
{Arabidopsis thaliana}, partial (44%)
Length = 921
Score = 134 bits (336), Expect = 2e-31
Identities = 81/228 (35%), Positives = 127/228 (55%), Gaps = 11/228 (4%)
Frame = +2
Query: 458 LMQEVHLLSRLRHPNIVQYYGSETVDDKLYIYLEYVSGGS-IHKLLQEYGQ-FGELAIRS 515
+ +EV +S + HPN+++ + S T L++ + ++SGGS +H + + + F E I +
Sbjct: 23 IRREVQTMSLIDHPNLLRAHCSFTAGHSLWVVMPFMSGGSCLHIMKSSFPEGFDEPVIAT 202
Query: 516 YTQQILSGLAYLHAKNTLHRDIKGANILVDPNGRVKVADFGMAKHI--TG--QYCPLSFK 571
+++L L YLHA +HRD+K NIL+D NG VK+ADFG++ + TG Q +F
Sbjct: 203 VLREVLKALVYLHAHGHIHRDVKAGNILLDANGSVKMADFGVSACMFDTGDRQRSRNTFV 382
Query: 572 GSPYWMAPEVIKNSKECSLGVDIWSLGCTVLEMATTKPPWSQYEGVAAMFKIGNSKELPT 631
G+P WMAPEV++ DIWS G T LE+A P+S+Y + + + + P
Sbjct: 383 GTPCWMAPEVMQQLHGYDFKADIWSFGITALELAHGHAPFSKYPPMKVL--LMTLQNAPP 556
Query: 632 IPDH-----LSNEGKDFVRKCLQRNPRDRPSASELLDHPFVKGAAPLE 674
D+ S K+ V CL ++P+ RPS+ +LL H F K A E
Sbjct: 557 GLDYERDKRFSKSFKELVATCLVKDPKKRPSSEKLLKHHFFKHARATE 700
>TC86454 homologue to GP|4567091|gb|AAD23582.1| SNF-1-like serine/threonine
protein kinase {Glycine max}, partial (90%)
Length = 2194
Score = 133 bits (335), Expect = 2e-31
Identities = 92/307 (29%), Positives = 148/307 (47%), Gaps = 8/307 (2%)
Frame = +3
Query: 382 FSHSNSAATSPSMPRSPARADSPMSSGS--------RWKKGKLLGRGTFGHVYIGFNSQS 433
F HS SA + S+ + D G +K GK LG G+FG V I + +
Sbjct: 3 FLHSLSATSIFSL*SLILKMDGSAGPGGGNVNAFLRNYKMGKTLGIGSFGKVKIAEHVLT 182
Query: 434 GEMCAMKEVTLFSDDAKSLESAKQLMQEVHLLSRLRHPNIVQYYGSETVDDKLYIYLEYV 493
G A+K L K++E +++ +E+ +L H +I++ Y +Y+ +EYV
Sbjct: 183 GHKVAIK--ILNRRKIKNMEMEEKVRREIKILRLFMHHHIIRLYEVVETPTDIYVVMEYV 356
Query: 494 SGGSIHKLLQEYGQFGELAIRSYTQQILSGLAYLHAKNTLHRDIKGANILVDPNGRVKVA 553
G + + E G+ E RS+ QQI+SG+ Y H +HRD+K N+L+D VK+A
Sbjct: 357 KSGELFDYIVEKGRLQEDEARSFFQQIISGVEYCHRNMVVHRDLKPENLLLDSKWSVKIA 536
Query: 554 DFGMAKHITGQYCPLSFKGSPYWMAPEVIKNSKECSLGVDIWSLGCTVLEMATTKPPWSQ 613
DFG++ + + + GSP + APEVI VD+WS G + + P+
Sbjct: 537 DFGLSNIMRDGHFLKTSCGSPNYAAPEVISGKLYAGPEVDVWSCGVILYALLCGTLPFDD 716
Query: 614 YEGVAAMFKIGNSKELPTIPDHLSNEGKDFVRKCLQRNPRDRPSASELLDHPFVKGAAPL 673
E + +FK + T+P HLS +D + + L +P R + E+ HP+ + L
Sbjct: 717 -ENIPNLFK-KIKGGIYTLPSHLSPGARDLIPRLLVVDPMKRMTIPEIRQHPWFQ--LHL 884
Query: 674 ERPIMVP 680
R + VP
Sbjct: 885 PRYLAVP 905
>TC77782 similar to GP|13448037|gb|AAK26845.1 SOS2-like protein kinase PKS6
{Arabidopsis thaliana}, partial (77%)
Length = 1236
Score = 127 bits (320), Expect = 1e-29
Identities = 81/271 (29%), Positives = 136/271 (49%), Gaps = 2/271 (0%)
Frame = +2
Query: 404 PMSSGSRWKKGKLLGRGTFGHVYIGFNSQSGEMCAMKEVTLFSDDAKSLESAKQLMQEVH 463
P + +++ GK +G G+F V + N ++G+ A+K L + QL +E+
Sbjct: 170 PRTRVGKYEMGKTIGEGSFAKVKLAKNVENGDYVAIK--ILDRNHVLKHNMMDQLKREIS 343
Query: 464 LLSRLRHPNIVQYYGSETVDDKLYIYLEYVSGGSIHKLLQEYGQFGELAIRSYTQQILSG 523
+ + HPN+++ + K+YI LE V+GG + + +G+ E RSY QQ+++
Sbjct: 344 AMKIINHPNVIKIFEVMASKTKIYIVLELVNGGELFDKIATHGRLKEDEARSYFQQLINA 523
Query: 524 LAYLHAKNTLHRDIKGANILVDPNGRVKVADFGMAKHITGQYCPL-SFKGSPYWMAPEVI 582
+ Y H++ HRD+K N+L+D NG +KV+DFG++ + + L + G+P ++APEV+
Sbjct: 524 VDYCHSRGVYHRDLKPENLLLDTNGVLKVSDFGLSTYSQQEDELLRTACGTPNYVAPEVL 703
Query: 583 KNSKECSLGVDIWSLGCTVLEMATTKPPWSQYEGVAAMFKIGNSKELPTIPDHLSNEGKD 642
+ DIWS G + + P+ + +A KIG + P S E K
Sbjct: 704 NDRGYVGSSSDIWSCGVILFVLMAGYLPFDEPNLIALYRKIGRADF--KCPSWFSPEAKK 877
Query: 643 FVRKCLQRNPRDRPSASELL-DHPFVKGAAP 672
+ L NP R E+L D F KG P
Sbjct: 878 LLTSILNPNPLTRIKIPEILEDEWFRKGYKP 970
>TC78699 weakly similar to PIR|G96761|G96761 probable MAP kinase T9L24.32
[imported] - Arabidopsis thaliana, partial (71%)
Length = 1119
Score = 127 bits (319), Expect = 2e-29
Identities = 104/343 (30%), Positives = 171/343 (49%), Gaps = 15/343 (4%)
Frame = +3
Query: 340 VTPIHPRAGG---TPTESQTGRADDGKQQSHRLPLP-PLTVTNTSPFSHSNSAATSPSMP 395
+T + P G T S R + R P+P P + + +++AT+ S+
Sbjct: 9 ITTVQPPVYGRLSTCRNSPKLRLPEISDHRPRFPVPLPANI-----YKQPSTSATTASV- 170
Query: 396 RSPARADSPMSSGSRWKKGKLLGRGTFGHVYIGFNSQSGEMCAMKEVTLFSDDAKSLESA 455
A S ++K +LG G G VY + + + A+K SD +
Sbjct: 171 -----AGGDNISAGDFEKLSVLGHGNGGTVYKVRHKLTSIIYALKINHYDSDPT----TR 323
Query: 456 KQLMQEVHLLSRLRH-PNIVQYYGS-ETVDDKLYIYLEYVSGGSIHKLLQEYGQFGELAI 513
++ + EV++L R N+V+Y+GS E + I +EY+ GS+ L+ G F E +
Sbjct: 324 RRALTEVNILRRATDCTNVVKYHGSFEKPTGDVCILMEYMDSGSLETALKTTGTFSESKL 503
Query: 514 RSYTQQILSGLAYLHAKNTLHRDIKGANILVDPNGRVKVADFGMAKHI--TGQYCPLSFK 571
+ + IL+GL YLHA+N HRDIK +NILV+ VK+ADFG++K + T + C S+
Sbjct: 504 STVARDILNGLTYLHARNIAHRDIKPSNILVNIKNEVKIADFGVSKFMGRTLEACN-SYV 680
Query: 572 GSPYWMAPE----VIKNSKECSLGVDIWSLGCTVLEMATTKPPW---SQYEGVAAMFKIG 624
G+ +M+PE + DIWSLG T+ E+ P+ Q A++
Sbjct: 681 GTCAYMSPERFDPEVYGGNYNGFSADIWSLGLTLFELYVGYFPFLQSGQRPDWASLMCAI 860
Query: 625 NSKELPTIPDHLSNEGKDFVRKCLQRNPRDRPSASELLDHPFV 667
+ P++P+ S+E ++FV CL++ +R SA++LL HPF+
Sbjct: 861 CFSDPPSLPETASSEFRNFVECCLKKESGERWSAAQLLTHPFL 989
>TC79461 similar to GP|9758254|dbj|BAB08753.1 contains similarity to
NRK-related kinase~gene_id:MWC10.1 {Arabidopsis
thaliana}, partial (88%)
Length = 1230
Score = 127 bits (318), Expect = 2e-29
Identities = 87/260 (33%), Positives = 145/260 (55%), Gaps = 8/260 (3%)
Frame = +1
Query: 415 KLLGRGTFGHVYIGFNSQSGEMCAMKEVTL--FSDDAKSLESAKQLMQEVHLLSRLRHPN 472
+LLG G VY F+ + G A +V L F D+ +E +L EV LL L + N
Sbjct: 136 ELLGCGAVKKVYRAFDQEEGIEVAWNQVKLRNFCDEPAMVE---RLYSEVRLLRSLTNKN 306
Query: 473 IVQYYG--SETVDDKLYIYLEYVSGGSIHKLLQEYGQFGELAIRSYTQQILSGLAYLHAK 530
I++ Y S+ ++ L E + G++ + +++ A++ +++QIL GL YLH
Sbjct: 307 IIELYSVWSDDRNNTLNFITEVCTSGNLREYRKKHRHVSMKALKKWSRQILKGLNYLHTH 486
Query: 531 NT--LHRDIKGANILVDPN-GRVKVADFGMAKHITGQYCPLSFKGSPYWMAPEVIKNSKE 587
+HRD+ +N+ V+ N G+VK+ D G+A + + + G+P +MAPE+ ++
Sbjct: 487 EPCIIHRDLNCSNVFVNGNVGQVKIGDLGLAAIVGKNHIAHTILGTPEFMAPELY--DED 660
Query: 588 CSLGVDIWSLGCTVLEMATTKPPWSQYEGVAAMFKIGNSKELPTIPDHL-SNEGKDFVRK 646
+ VDI+S G VLEM T + P+S+ + VA ++K +S P + + +E K+F+ +
Sbjct: 661 YTELVDIYSFGMCVLEMVTLEIPYSECDNVAKIYKKVSSGIRPAAMNKVKDSEVKEFIER 840
Query: 647 CLQRNPRDRPSASELLDHPF 666
CL + PR RPSA+ELL PF
Sbjct: 841 CLAQ-PRARPSAAELLKDPF 897
>TC79149 similar to PIR|T46059|T46059 MAP kinase - Arabidopsis thaliana,
partial (63%)
Length = 1671
Score = 127 bits (318), Expect = 2e-29
Identities = 90/267 (33%), Positives = 151/267 (55%), Gaps = 6/267 (2%)
Frame = +3
Query: 407 SGSRWKKGKLLGRGTFGHVYIGFNSQSGEMCAMKEVTLFSDDAKSLESAKQLMQEVHLLS 466
+G + G +LG+G VY + G A +V L ++ + + ++L EVHLLS
Sbjct: 408 TGRYGRFGDVLGKGAMKTVYKAIDEVLGIEVAWNQVRL-NEVLNTPDDLQRLYSEVHLLS 584
Query: 467 RLRHPNIVQYYGSET-VDDKLYIYL-EYVSGGSIHKLLQEYGQFGELAIRSYTQQILSGL 524
L+H +I+++Y S +D+K + ++ E + GS+ + ++Y + AI+S+ +QIL GL
Sbjct: 585 TLKHRSIMRFYTSWIDIDNKNFNFVTEMFTSGSLREYRRKYKRVSLQAIKSWARQILQGL 764
Query: 525 AYLHAKN--TLHRDIKGANILVDPN-GRVKVADFGMAKHITGQYCPLSFKGSPYWMAPEV 581
YLH + +HRD K NI V+ + G+VK+ D G+A + G S G+P +MAPE+
Sbjct: 765 VYLHGHDPPVIHRDPKCDNIFVNGHLGQVKIGDLGLAAILQGSQSAHSVIGTPEFMAPEM 944
Query: 582 IKNSKECSLGVDIWSLGCTVLEMATTKPPWSQYEGVAAMFKIGNSKELPTIPDHLSN-EG 640
+ +E + D++S G VLEM T+ P+S+ A ++K S +LP + + E
Sbjct: 945 YE--EEYNELADVYSFGMCVLEMLTSDYPYSECTNPAQIYKKVTSGKLPMSFFRIEDGEA 1118
Query: 641 KDFVRKCLQRNPRDRPSASELLDHPFV 667
+ F+ KCL+ +RPSA +LL PF+
Sbjct: 1119RRFIGKCLE-PAANRPSAKDLLLEPFL 1196
>TC77210 similar to GP|21954717|gb|AAM83095.1 SOS2-like protein kinase
{Glycine max}, partial (88%)
Length = 1831
Score = 126 bits (317), Expect = 3e-29
Identities = 82/269 (30%), Positives = 140/269 (51%), Gaps = 6/269 (2%)
Frame = +1
Query: 410 RWKKGKLLGRGTFGHVYIGFNSQSGEMCAMKEVTLFSDDAKSLESAKQLMQEVHLLSRLR 469
+++ G++LG GTF VY N +G+ AMK V + + +Q+ +E+ ++ ++
Sbjct: 352 KYELGRVLGHGTFAKVYHAKNLNTGKNVAMKVVG--KEKVIKVGMIEQIKREISVMKMVK 525
Query: 470 HPNIVQYYGSETVDDKLYIYLEYVSGGSIHKLLQEYGQFGELAIRSYTQQILSGLAYLHA 529
HPNIVQ + K+YI +E V GG + + + G+ E R Y QQ++S + + H+
Sbjct: 526 HPNIVQLHEVMASKSKIYIAMELVRGGELFNKIVK-GRLKEDVARVYFQQLISAVDFCHS 702
Query: 530 KNTLHRDIKGANILVDPNGRVKVADFGM---AKHITGQYCPLSFKGSPYWMAPEVIKNSK 586
+ HRD+K N+L+D +G +KV+DFG+ ++H+ + G+P +++PEVI
Sbjct: 703 RGVYHRDLKPENLLLDEDGNLKVSDFGLCTFSEHLRQDGLLHTTCGTPAYVSPEVIAKKG 882
Query: 587 ECSLGVDIWSLGCTVLEMATTKPPWSQYEGVAAMFK---IGNSKELPTIPDHLSNEGKDF 643
DIWS G + + P+ Q + + AM+K G+ K P S E +
Sbjct: 883 YDGAKADIWSCGVILYVLLAGFLPF-QDDNLVAMYKKIYRGDFK----CPPWFSLEARRL 1047
Query: 644 VRKCLQRNPRDRPSASELLDHPFVKGAAP 672
+ K L NP R S S+++D + K P
Sbjct: 1048ITKLLDPNPNTRISISKIMDSSWFKKPVP 1134
>TC77881 homologue to GP|15528439|emb|CAC69137. MEK map kinase kinsae
{Medicago sativa subsp. x varia}, complete
Length = 1546
Score = 125 bits (313), Expect = 8e-29
Identities = 107/364 (29%), Positives = 180/364 (49%), Gaps = 13/364 (3%)
Frame = +2
Query: 317 EYSPIPSPRMTSPGPSSRIQSGAVTPIHPRAGGTPTESQTGRADDGKQQSHRLPLPP--- 373
E PI P TS G ++ A G+P + + R ++ LPLP
Sbjct: 215 EMRPIQLPPPTSTGSAT-------------ASGSPNNNNS-RPQRRRRDHLTLPLPQRDT 352
Query: 374 -LTVTNTSPFSHSNSAATSPSMPRSPARADSPMSSGSRWKKGKLLGRGTFGHVYIGFNSQ 432
L V P S + + S S + + S ++ +G G+ G VY +
Sbjct: 353 NLAVPLPLPPSGGSGGGGNGSGSGSGGASQQLVIPFSELERLNRIGSGSGGTVYKVVHRI 532
Query: 433 SGEMCAMKEVTLFSDDAKSLESAKQLMQEVHLLSRLRHPNIVQYYGSETVDDKLYIYLEY 492
+G A+K ++ +S+ +Q+ +E+ +L + N+V+ + + ++ + LEY
Sbjct: 533 NGRAYALK--VIYGHHEESVR--RQIHREIQILRDVDDVNVVKCHEMYDHNAEIQVLLEY 700
Query: 493 VSGGSIH-KLLQEYGQFGELAIRSYTQQILSGLAYLHAKNTLHRDIKGANILVDPNGRVK 551
+ GGS+ K + + Q ++A +QIL GLAYLH ++ +HRDIK +N+L++ +VK
Sbjct: 701 MDGGSLEGKHIPQENQLADVA-----RQILRGLAYLHRRHIVHRDIKPSNLLINSRKQVK 865
Query: 552 VADFGMAKHITGQYCPL-SFKGSPYWMAPE----VIKNSKECSLGVDIWSLGCTVLEMAT 606
+ADFG+ + + P S G+ +M+PE I + + + DIWSLG ++LE
Sbjct: 866 IADFGVGRILNQTMDPCNSSVGTIAYMSPERINTDINDGQYDAYAGDIWSLGVSILEFYM 1045
Query: 607 TKPPWS---QYEGVAAMFKIGNSKELPTIPDHLSNEGKDFVRKCLQRNPRDRPSASELLD 663
+ P++ Q + + M I S+ P P S E +DFV +CLQR+P R +AS LL
Sbjct: 1046GRFPFAVGRQGDWASLMCAICMSQP-PEAPTTASPEFRDFVSRCLQRDPSRRWTASRLLS 1222
Query: 664 HPFV 667
HPF+
Sbjct: 1223HPFL 1234
>TC77429 similar to GP|15215664|gb|AAK91377.1 AT5g25110/T11H3_120
{Arabidopsis thaliana}, partial (79%)
Length = 1869
Score = 124 bits (312), Expect = 1e-28
Identities = 80/264 (30%), Positives = 136/264 (51%), Gaps = 4/264 (1%)
Frame = +3
Query: 410 RWKKGKLLGRGTFGHVYIGFNSQSGEMCAMKEVTLFSDDAKSLESAKQLMQEVHLLSRLR 469
+++ G++LG+GTF VY +GE A+K V D K +Q+ +E+ ++ ++
Sbjct: 267 KYEMGRVLGKGTFAKVYYAKEISTGEGVAIKVVD--KDKVKKEGMMEQIKREISVMRLVK 440
Query: 470 HPNIVQYYGSETVDDKLYIYLEYVSGGSIHKLLQEYGQFGELAIRSYTQQILSGLAYLHA 529
HPNIV K+ +EY GG + + + + G+F E R Y QQ++S + Y H+
Sbjct: 441 HPNIVNLKEVMATKTKILFVMEYARGGELFEKVAK-GKFKEELARKYFQQLISAVDYCHS 617
Query: 530 KNTLHRDIKGANILVDPNGRVKVADFGMA---KHITGQYCPLSFKGSPYWMAPEVIKNSK 586
+ HRD+K N+L+D N +KV+DFG++ +H+ + G+P ++APEV++
Sbjct: 618 RGVSHRDLKPENLLLDENENLKVSDFGLSALPEHLRQDGLLHTQCGTPAYVAPEVVRKRG 797
Query: 587 ECSLGVDIWSLGCTVLEMATTKPPWSQYEGVAAMFKIGNSKELPTIPDHLSNEGKDFVRK 646
D WS G + + P+ Q+E + +M+ KE P S E K + K
Sbjct: 798 YSGFKADTWSCGVILYALLAGFLPF-QHENLISMYN-KVFKEEYQFPPWFSPESKRLISK 971
Query: 647 CLQRNPRDRPSASELLDHP-FVKG 669
L +P R + S +++ P F KG
Sbjct: 972 ILVADPERRITISSIMNVPWFQKG 1043
>TC90430 similar to GP|18766642|gb|AAL79042.1 NIMA-related protein kinase
{Populus x canescens}, partial (45%)
Length = 976
Score = 123 bits (308), Expect = 3e-28
Identities = 79/234 (33%), Positives = 130/234 (54%), Gaps = 4/234 (1%)
Frame = +1
Query: 439 MKEVTLFSDDAKSLESAKQLMQEVHLLSRLRHPNIVQYYGSETVDDKLY--IYLEYVSGG 496
+K++ L ++ SA Q E+ L+S++R+P IV+Y S V+ + I + Y GG
Sbjct: 169 LKKIRLARQTDRTRRSAHQ---EMELISKVRNPFIVEYKDS-WVEKGCFVCIIIGYCEGG 336
Query: 497 SIHKLLQEYG--QFGELAIRSYTQQILSGLAYLHAKNTLHRDIKGANILVDPNGRVKVAD 554
+ + +++ F E + + Q+L L YLH + LHRD+K +NI + N +++ D
Sbjct: 337 DMAETVKKANGVNFPEEKLCKWLVQLLMALDYLHVNHILHRDVKCSNIFLTKNQDIRLGD 516
Query: 555 FGMAKHITGQYCPLSFKGSPYWMAPEVIKNSKECSLGVDIWSLGCTVLEMATTKPPWSQY 614
FG+AK +T S G+P +M PE++ + S DIWSLGC V EMA +P + +
Sbjct: 517 FGLAKLLTSDDLASSIVGTPSYMCPELLADIPYGSKS-DIWSLGCCVYEMAAHRPAFKAF 693
Query: 615 EGVAAMFKIGNSKELPTIPDHLSNEGKDFVRKCLQRNPRDRPSASELLDHPFVK 668
+ A + KI S P +P S+ + V+ L++NP RP+A ELL+HP ++
Sbjct: 694 DIQALIHKINKSIVSP-LPTMYSSAFRGLVKSMLRKNPELRPTAGELLNHPHLQ 852
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.313 0.131 0.389
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 24,416,509
Number of Sequences: 36976
Number of extensions: 391273
Number of successful extensions: 3436
Number of sequences better than 10.0: 623
Number of HSP's better than 10.0 without gapping: 2922
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 3084
length of query: 754
length of database: 9,014,727
effective HSP length: 103
effective length of query: 651
effective length of database: 5,206,199
effective search space: 3389235549
effective search space used: 3389235549
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.9 bits)
S2: 62 (28.5 bits)
Medicago: description of AC121237.19


