

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC121233.14 - phase: 0
(275 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
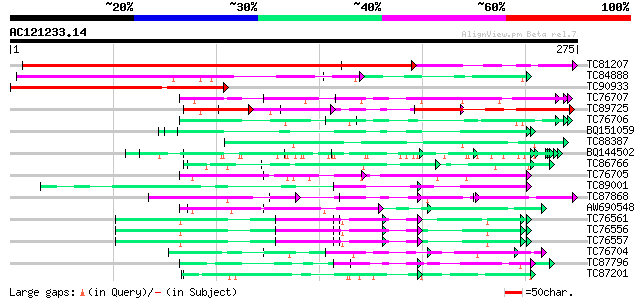
Score E
Sequences producing significant alignments: (bits) Value
TC81207 similar to GP|21322752|dbj|BAB78536. cold shock protein-... 419 e-118
TC84888 similar to GP|21322752|dbj|BAB78536. cold shock protein-... 139 2e-33
TC90933 similar to SP|P27484|GRP2_NICSY Glycine-rich protein 2. ... 119 1e-27
TC76707 similar to SP|Q07202|CORA_MEDSA Cold and drought-regulat... 89 2e-18
TC89725 similar to PIR|T05494|T05494 glycine-rich protein T19K4.... 85 3e-17
TC76706 similar to SP|Q07202|CORA_MEDSA Cold and drought-regulat... 69 1e-12
BQ151059 similar to GP|7291525|gb|A CG9888-PA {Drosophila melano... 68 4e-12
TC88387 similar to GP|13357253|gb|AAK20050.1 putative zinc finge... 65 2e-11
BQ144502 weakly similar to GP|6523547|emb hydroxyproline-rich gl... 64 5e-11
TC86766 weakly similar to PIR|T06291|T06291 extensin homolog T9E... 64 5e-11
TC76705 similar to SP|Q07202|CORA_MEDSA Cold and drought-regulat... 63 1e-10
TC89001 similar to GP|5726567|gb|AAD48471.1| glycine-rich RNA-bi... 62 2e-10
TC87868 similar to PIR|T05112|T05112 splicing factor 9G8-like SR... 60 7e-10
AW690548 similar to SP|Q09134|GRPA Abscisic acid and environment... 58 3e-09
TC76561 similar to PIR|S71779|S71779 glycine-rich RNA-binding pr... 54 5e-08
TC76556 similar to PIR|S71779|S71779 glycine-rich RNA-binding pr... 54 5e-08
TC76557 similar to PIR|S71779|S71779 glycine-rich RNA-binding pr... 54 5e-08
TC76704 homologue to SP|Q09134|GRPA_MEDFA Abscisic acid and envi... 54 8e-08
TC87796 weakly similar to SP|P03211|EBN1_EBV EBNA-1 nuclear prot... 54 8e-08
TC87201 similar to PIR|F86388|F86388 hypothetical protein AAG292... 53 1e-07
>TC81207 similar to GP|21322752|dbj|BAB78536. cold shock protein-1 {Triticum
aestivum}, partial (39%)
Length = 630
Score = 419 bits (1078), Expect = e-118
Identities = 190/191 (99%), Positives = 191/191 (99%)
Frame = +3
Query: 7 SGVVQWFNNTKGFGFIKPDDDSEDLFVHQSDIRSEGFRSLSEGDRVEFTIADGDNGKTKA 66
+GVVQWFNNTKGFGFIKPDDDSEDLFVHQSDIRSEGFRSLSEGDRVEFTIADGDNGKTKA
Sbjct: 57 AGVVQWFNNTKGFGFIKPDDDSEDLFVHQSDIRSEGFRSLSEGDRVEFTIADGDNGKTKA 236
Query: 67 VDVTGPKGEPLQVRQDNHGGGGGGRGFRGGERRNGGGGCYTCGDTGHIARDCDRSDRNDR 126
VDVTGPKGEPLQVRQDNHGGGGGGRGFRGGERRNGGGGCYTCGDTGHIARDCDRSDRNDR
Sbjct: 237 VDVTGPKGEPLQVRQDNHGGGGGGRGFRGGERRNGGGGCYTCGDTGHIARDCDRSDRNDR 416
Query: 127 NDRSGGGGGGDRDRACYTCGSFEHFARDCMRGGGNNNNGGGGYGGGGTSCYRCGGVGHIA 186
NDRSGGGGGGDRDRACYTCGSFEHFARDCMRGGGNNNNGGGGYGGGGTSCYRCGGVGHIA
Sbjct: 417 NDRSGGGGGGDRDRACYTCGSFEHFARDCMRGGGNNNNGGGGYGGGGTSCYRCGGVGHIA 596
Query: 187 RDCATPSSGGG 197
RDCATPSSGGG
Sbjct: 597 RDCATPSSGGG 629
Score = 68.2 bits (165), Expect = 3e-12
Identities = 43/114 (37%), Positives = 50/114 (43%)
Frame = +3
Query: 162 NNNGGGGYGGGGTSCYRCGGVGHIARDCATPSSGGGGGGACYKCGEVGHIARDCSNEGGR 221
+N+GGGG G G +R G GGG CY CG+ GHIARDC
Sbjct: 282 DNHGGGGGGRG----FRGG-------------ERRNGGGGCYTCGDTGHIARDCDRSDR- 407
Query: 222 FDGGNGRYDDGNGRFGGGNRRFGSGGGGHDGGKGTCFNCGKAGHFARDCVEASG 275
+D N R GGG GGG D C+ CG HFARDC+ G
Sbjct: 408 --------NDRNDRSGGG------GGGDRDRA---CYTCGSFEHFARDCMRGGG 518
>TC84888 similar to GP|21322752|dbj|BAB78536. cold shock protein-1 {Triticum
aestivum}, partial (58%)
Length = 612
Score = 139 bits (349), Expect = 2e-33
Identities = 83/180 (46%), Positives = 101/180 (56%), Gaps = 11/180 (6%)
Frame = +2
Query: 4 ERISGVVQWFNNTKGFGFIKPDDDSEDLFVHQSDIRSEGFRSLSEGDRVEFTIADGDNGK 63
E+ +G V+WFN+TKGFGFI PD EDLFVHQ+ I+++GFRSL+EG+ VEF + GD+G+
Sbjct: 53 EKQTGTVKWFNSTKGFGFISPDGGGEDLFVHQTSIKTDGFRSLAEGEAVEFIVEAGDDGR 232
Query: 64 TKAVDVTGPKGEPLQ--VRQDNHGGGGGGR---GFRGG--ERRNGGGGCYTCGDTGHIAR 116
TKA+DVTGP G P Q RQ+ GGGGGR G+ GG + GGGG Y G
Sbjct: 233 TKAIDVTGPNGAPPQGAPRQEMGYGGGGGRGGGGYGGGYDQGYGGGGGSYGGG------- 391
Query: 117 DCDRSDRNDRNDRSGGGGGGDRDRACYTCGSFEHFARDCMRGGGNNNNGG----GGYGGG 172
GGG GG R Y G + GGG GG GGYGGG
Sbjct: 392 -------------FGGGRGGGRGGGGYXQGGY--------GGGGGYXQGGYGGQGGYGGG 508
Score = 45.4 bits (106), Expect = 2e-05
Identities = 38/102 (37%), Positives = 40/102 (38%), Gaps = 1/102 (0%)
Frame = +2
Query: 153 RDCMRGGGNNNNGGGGYGGGGTSCYRCGGVGHIARDCATPSSGGGGGGACYKCGEVGHIA 212
R M GG GGGGYGGG Y GG S GGG GG
Sbjct: 287 RQEMGYGGGGGRGGGGYGGGYDQGYGGGG----------GSYGGGFGG------------ 400
Query: 213 RDCSNEGGRFDGGNGRYDDGNGRFGGGNRRFGSGG-GGHDGG 253
GGR G G Y G GGG + G GG GG+ GG
Sbjct: 401 ---GRGGGR---GGGGYXQGGYGGGGGYXQGGYGGQGGYGGG 508
Score = 35.4 bits (80), Expect = 0.024
Identities = 20/39 (51%), Positives = 23/39 (58%), Gaps = 2/39 (5%)
Frame = +2
Query: 219 GGRFDGG-NGRYDDGNGRFGGGNRRFGSG-GGGHDGGKG 255
GGR GG G YD G +GGG +G G GGG GG+G
Sbjct: 314 GGRGGGGYGGGYDQG---YGGGGGSYGGGFGGGRGGGRG 421
Score = 33.9 bits (76), Expect = 0.069
Identities = 31/89 (34%), Positives = 33/89 (36%), Gaps = 1/89 (1%)
Frame = +2
Query: 168 GYGGGGTSCYRCGGVGHIARDCATPSSGGGGGGACYKCGEVGHIARDCSNEGGRFDGGNG 227
GYGGGG GGGG G Y G + GG G
Sbjct: 299 GYGGGG-------------------GRGGGGYGGGYDQG---------------YGGGGG 376
Query: 228 RYDDGNGRFGGGNRRFGSGGGGH-DGGKG 255
Y G FGGG R G GGGG+ GG G
Sbjct: 377 SYGGG---FGGG-RGGGRGGGGYXQGGYG 451
>TC90933 similar to SP|P27484|GRP2_NICSY Glycine-rich protein 2. [Wood
tobacco] {Nicotiana sylvestris}, partial (44%)
Length = 368
Score = 119 bits (299), Expect = 1e-27
Identities = 58/106 (54%), Positives = 76/106 (70%)
Frame = +3
Query: 1 MAEERISGVVQWFNNTKGFGFIKPDDDSEDLFVHQSDIRSEGFRSLSEGDRVEFTIADGD 60
M+ +++G V+WFN+ KGFGFI PDD SE+LFVHQS I+++GFRSL+EG+ VE+ I +
Sbjct: 57 MSGSKLTGKVKWFNDQKGFGFITPDDGSEELFVHQSQIQTDGFRSLAEGESVEYQIESDN 236
Query: 61 NGKTKAVDVTGPKGEPLQVRQDNHGGGGGGRGFRGGERRNGGGGCY 106
+G++KAV VTGP G V+ GGGGGG RGG GGGG Y
Sbjct: 237 DGRSKAVSVTGPDG--ASVQGSRRGGGGGGGYERGGGGYGGGGGGY 368
Score = 30.4 bits (67), Expect = 0.77
Identities = 14/32 (43%), Positives = 18/32 (55%)
Frame = +3
Query: 224 GGNGRYDDGNGRFGGGNRRFGSGGGGHDGGKG 255
G +G G+ R GGG + GGGG+ GG G
Sbjct: 267 GPDGASVQGSRRGGGGGGGYERGGGGYGGGGG 362
>TC76707 similar to SP|Q07202|CORA_MEDSA Cold and drought-regulated protein
CORA. [Alfalfa] {Medicago sativa}, partial (87%)
Length = 780
Score = 89.0 bits (219), Expect = 2e-18
Identities = 67/194 (34%), Positives = 81/194 (41%), Gaps = 9/194 (4%)
Frame = +1
Query: 83 NHGGGG---GGRGFRGGERRNGGGGCYTCGDTGHIARDCDRSDRNDRNDRSGGGGGGDRD 139
NHGGGG GG GG NGGGG Y G G+ + GGGG +
Sbjct: 178 NHGGGGYNGGGYNHGGGGYNNGGGG-YNHGGGGY----------------NNGGGGYNHG 306
Query: 140 RACYTCGSFEHFARDCMRGGGNNNNGGGGYGGGGTSCYRCGGVGHIARDCATPSSGGGGG 199
Y G + H GGG N+GGGGY GGG GG G+ +GGGG
Sbjct: 307 GGGYNGGGYNHGGGGYNHGGGGYNHGGGGYNGGGGHGGHGGG-GY---------NGGGGH 456
Query: 200 GACYKCGEVGHIARDCSNE------GGRFDGGNGRYDDGNGRFGGGNRRFGSGGGGHDGG 253
G V + +NE GG ++ G G Y+ G G + G + GGGGH G
Sbjct: 457 GGHGAAESVAVQTEEKTNEVNDAKYGGGYNHGGGSYNHGGGSYHHGGGGYNHGGGGHGGH 636
Query: 254 KGTCFNCGKAGHFA 267
G G GH A
Sbjct: 637 GGG----GHGGHGA 666
Score = 73.6 bits (179), Expect = 8e-14
Identities = 56/152 (36%), Positives = 68/152 (43%), Gaps = 4/152 (2%)
Frame = +1
Query: 124 NDRNDRSGGGGGGDRDRAC--YTCGSFEHFARDCMRGGGNNNNGGGGY--GGGGTSCYRC 179
N+ ND GG GG + Y G + H GGG NNGGGGY GGGG Y
Sbjct: 133 NEVNDAKYGGYGGGYNHGGGGYNGGGYNH-------GGGGYNNGGGGYNHGGGG---YNN 282
Query: 180 GGVGHIARDCATPSSGGGGGGACYKCGEVGHIARDCSNEGGRFDGGNGRYDDGNGRFGGG 239
GG G+ + GGGG Y G H GG ++ G G Y+ G G + GG
Sbjct: 283 GGGGY--------NHGGGG----YNGGGYNH-------GGGGYNHGGGGYNHGGGGYNGG 405
Query: 240 NRRFGSGGGGHDGGKGTCFNCGKAGHFARDCV 271
G GGGG++GG G GH A + V
Sbjct: 406 GGHGGHGGGGYNGGG------GHGGHGAAESV 483
Score = 62.4 bits (150), Expect = 2e-10
Identities = 45/120 (37%), Positives = 55/120 (45%), Gaps = 5/120 (4%)
Frame = +1
Query: 159 GGNNNNGGGGYGGGGTSCYRCGGVGHIARDCATPSSGGGG---GGACYKCGEVGHIARDC 215
GG N+GGGGY GGG Y GG G+ ++GGGG GG Y G G+
Sbjct: 166 GGGYNHGGGGYNGGG---YNHGGGGY--------NNGGGGYNHGGGGYNNGGGGYNHGGG 312
Query: 216 SNEGGRFDGGNGRYDDGNGRF--GGGNRRFGSGGGGHDGGKGTCFNCGKAGHFARDCVEA 273
GG ++ G G Y+ G G + GGG G GGG GG G G GH E+
Sbjct: 313 GYNGGGYNHGGGGYNHGGGGYNHGGG----GYNGGGGHGGHGGGGYNGGGGHGGHGAAES 480
Score = 29.6 bits (65), Expect = 1.3
Identities = 17/50 (34%), Positives = 23/50 (46%), Gaps = 5/50 (10%)
Frame = +1
Query: 83 NHGGG-----GGGRGFRGGERRNGGGGCYTCGDTGHIARDCDRSDRNDRN 127
NHGGG GG GG +GGGG G GH ++++ +N
Sbjct: 544 NHGGGSYNHGGGSYHHGGGGYNHGGGGHGGHGGGGHGGHGAEQTEDKTQN 693
>TC89725 similar to PIR|T05494|T05494 glycine-rich protein T19K4.150 -
Arabidopsis thaliana, partial (17%)
Length = 378
Score = 85.1 bits (209), Expect = 3e-17
Identities = 39/78 (50%), Positives = 49/78 (62%)
Frame = +1
Query: 197 GGGGACYKCGEVGHIARDCSNEGGRFDGGNGRYDDGNGRFGGGNRRFGSGGGGHDGGKGT 256
G GG CY CGE GH+AR+C++ GG G GR+GGG GGGG GG G+
Sbjct: 1 GVGGGCYNCGESGHMARECTSGGG----------GGGGRYGGGG-----GGGGGGGGGGS 135
Query: 257 CFNCGKAGHFARDCVEAS 274
C++CG++GHFARDC S
Sbjct: 136 CYSCGESGHFARDCPTGS 189
Score = 80.1 bits (196), Expect = 8e-16
Identities = 44/90 (48%), Positives = 46/90 (50%)
Frame = +1
Query: 132 GGGGGDRDRACYTCGSFEHFARDCMRGGGNNNNGGGGYGGGGTSCYRCGGVGHIARDCAT 191
G GGG CY CG H AR+C GGG GGG YGGGG GG G
Sbjct: 1 GVGGG-----CYNCGESGHMARECTSGGGG---GGGRYGGGG------GGGG-------- 114
Query: 192 PSSGGGGGGACYKCGEVGHIARDCSNEGGR 221
GGGGGG+CY CGE GH ARDC R
Sbjct: 115 ---GGGGGGSCYSCGESGHFARDCPTGSAR 195
Score = 67.0 bits (162), Expect = 7e-12
Identities = 29/60 (48%), Positives = 35/60 (58%), Gaps = 3/60 (5%)
Frame = +1
Query: 102 GGGCYTCGDTGHIARDCDRSDRND---RNDRSGGGGGGDRDRACYTCGSFEHFARDCMRG 158
GGGCY CG++GH+AR+C GGGGGG +CY+CG HFARDC G
Sbjct: 7 GGGCYNCGESGHMARECTSGGGGGGGRYGGGGGGGGGGGGGGSCYSCGESGHFARDCPTG 186
Score = 52.8 bits (125), Expect = 1e-07
Identities = 23/37 (62%), Positives = 26/37 (70%), Gaps = 3/37 (8%)
Frame = +1
Query: 85 GGGGGGR---GFRGGERRNGGGGCYTCGDTGHIARDC 118
GGGGGGR G GG GGG CY+CG++GH ARDC
Sbjct: 67 GGGGGGRYGGGGGGGGGGGGGGSCYSCGESGHFARDC 177
Score = 35.4 bits (80), Expect = 0.024
Identities = 12/24 (50%), Positives = 16/24 (66%)
Frame = +1
Query: 252 GGKGTCFNCGKAGHFARDCVEASG 275
G G C+NCG++GH AR+C G
Sbjct: 1 GVGGGCYNCGESGHMARECTSGGG 72
>TC76706 similar to SP|Q07202|CORA_MEDSA Cold and drought-regulated protein
CORA. [Alfalfa] {Medicago sativa}, partial (94%)
Length = 1034
Score = 69.3 bits (168), Expect = 1e-12
Identities = 57/187 (30%), Positives = 69/187 (36%), Gaps = 2/187 (1%)
Frame = +1
Query: 83 NHGGGGGGRGFRGGERRNGGGGCYTCGDTGHIARDCDRSDRNDRNDRSGG--GGGGDRDR 140
NHGGG G G+ G + GGG Y G G+ N GG GGGG
Sbjct: 211 NHGGGYNGGGYNHGGGYHNGGGGYHNGGGGY-------------NHGGGGYNGGGGHGGH 351
Query: 141 ACYTCGSFEHFARDCMRGGGNNNNGGGGYGGGGTSCYRCGGVGHIARDCATPSSGGGGGG 200
Y G GG NGGGG+GG G + V + + GG
Sbjct: 352 GGYNGG-----------GGHGGYNGGGGHGGHGAA----ESVAVQTEEKTNEVNDAKYGG 486
Query: 201 ACYKCGEVGHIARDCSNEGGRFDGGNGRYDDGNGRFGGGNRRFGSGGGGHDGGKGTCFNC 260
Y N GG ++ G G Y+ G G + G + GGGGH G G
Sbjct: 487 GSY-------------NHGGSYNHGGGSYNHGGGSYHHGGGGYNHGGGGHGGHGGG---- 615
Query: 261 GKAGHFA 267
G GH A
Sbjct: 616 GHGGHGA 636
Score = 44.3 bits (103), Expect = 5e-05
Identities = 36/106 (33%), Positives = 42/106 (38%), Gaps = 1/106 (0%)
Frame = +1
Query: 169 YGGGGTSCYRCGGVGHIARDCATPSSGGGGGGACYKCGEVGHIARDCSNEGGRFDGGNGR 228
YGG Y GG H GGG G Y G H N GG + G G
Sbjct: 184 YGG-----YNHGGYNH----------GGGYNGGGYNHGGGYH------NGGGGYHNGGGG 300
Query: 229 YDDGNGRFGGGNRRFGSGG-GGHDGGKGTCFNCGKAGHFARDCVEA 273
Y+ G G + GG G GG GG++GG G G GH E+
Sbjct: 301 YNHGGGGYNGGG---GHGGHGGYNGGGGHGGYNGGGGHGGHGAAES 429
Score = 42.7 bits (99), Expect = 1e-04
Identities = 41/124 (33%), Positives = 48/124 (38%), Gaps = 6/124 (4%)
Frame = +1
Query: 154 DCMRGGGNNN--NGGGGYGGGGTSCYRCGGVGHIARDCATPSSGGGGGGACYKCGEVGHI 211
D GG N+ N GGGY GGG Y GG H +GGGG
Sbjct: 175 DAKYGGYNHGGYNHGGGYNGGG---YNHGGGYH---------NGGGG------------- 279
Query: 212 ARDCSNEGGRFDGGNGRYDDGNGRFGGGNRRFG----SGGGGHDGGKGTCFNCGKAGHFA 267
+GG G Y+ G G + GG G +GGGGH G G G GH A
Sbjct: 280 ---------YHNGGGG-YNHGGGGYNGGGGHGGHGGYNGGGGHGGYNG---GGGHGGHGA 420
Query: 268 RDCV 271
+ V
Sbjct: 421 AESV 432
Score = 29.6 bits (65), Expect = 1.3
Identities = 17/50 (34%), Positives = 23/50 (46%), Gaps = 5/50 (10%)
Frame = +1
Query: 83 NHGGG-----GGGRGFRGGERRNGGGGCYTCGDTGHIARDCDRSDRNDRN 127
NHGGG GG GG +GGGG G GH ++++ +N
Sbjct: 514 NHGGGSYNHGGGSYHHGGGGYNHGGGGHGGHGGGGHGGHGAEQTEDKTQN 663
>BQ151059 similar to GP|7291525|gb|A CG9888-PA {Drosophila melanogaster},
partial (26%)
Length = 308
Score = 67.8 bits (164), Expect = 4e-12
Identities = 58/174 (33%), Positives = 59/174 (33%)
Frame = +2
Query: 82 DNHGGGGGGRGFRGGERRNGGGGCYTCGDTGHIARDCDRSDRNDRNDRSGGGGGGDRDRA 141
+N GGGGGG G GG GGGG GGGGGG
Sbjct: 26 ENPGGGGGGGGGGGGGGGGGGGG--------------------------GGGGGG----- 112
Query: 142 CYTCGSFEHFARDCMRGGGNNNNGGGGYGGGGTSCYRCGGVGHIARDCATPSSGGGGGGA 201
GGG GGGG GGGG GG G GGGGGG
Sbjct: 113 ----------------GGGGGGGGGGGGGGGG------GGGG---------GGGGGGGG- 196
Query: 202 CYKCGEVGHIARDCSNEGGRFDGGNGRYDDGNGRFGGGNRRFGSGGGGHDGGKG 255
GG GG G G G GGG G GGGG GG G
Sbjct: 197 -----------------GGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGG 307
Score = 65.1 bits (157), Expect = 3e-11
Identities = 58/180 (32%), Positives = 59/180 (32%)
Frame = +3
Query: 76 PLQVRQDNHGGGGGGRGFRGGERRNGGGGCYTCGDTGHIARDCDRSDRNDRNDRSGGGGG 135
P + GGGGGG G GG GGGG GGGGG
Sbjct: 15 PFPGKTPGGGGGGGGGGGGGGGGGGGGGG--------------------------GGGGG 116
Query: 136 GDRDRACYTCGSFEHFARDCMRGGGNNNNGGGGYGGGGTSCYRCGGVGHIARDCATPSSG 195
G GGG GGGG GGGG GG G G
Sbjct: 117 G---------------------GGGGGGGGGGGGGGGG------GGGG-----------G 182
Query: 196 GGGGGACYKCGEVGHIARDCSNEGGRFDGGNGRYDDGNGRFGGGNRRFGSGGGGHDGGKG 255
GGGGG GG GG G G G GGG G GGGG GG G
Sbjct: 183 GGGGG------------------GGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGG 308
Score = 60.8 bits (146), Expect = 5e-10
Identities = 55/181 (30%), Positives = 57/181 (31%)
Frame = +1
Query: 73 KGEPLQVRQDNHGGGGGGRGFRGGERRNGGGGCYTCGDTGHIARDCDRSDRNDRNDRSGG 132
+G+P GGGGGG G GG GGGG GG
Sbjct: 22 RGKPRGGGGGGGGGGGGGGGGGGGGGGGGGGG--------------------------GG 123
Query: 133 GGGGDRDRACYTCGSFEHFARDCMRGGGNNNNGGGGYGGGGTSCYRCGGVGHIARDCATP 192
GGGG GGG GGGG GGGG
Sbjct: 124 GGGG---------------------GGGGGGGGGGGGGGGG------------------- 183
Query: 193 SSGGGGGGACYKCGEVGHIARDCSNEGGRFDGGNGRYDDGNGRFGGGNRRFGSGGGGHDG 252
GGGGGG GG GG G G G GGG G GGGG G
Sbjct: 184 --GGGGGG------------------GGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGG 303
Query: 253 G 253
G
Sbjct: 304 G 306
Score = 34.7 bits (78), Expect = 0.041
Identities = 19/43 (44%), Positives = 20/43 (46%)
Frame = +3
Query: 213 RDCSNEGGRFDGGNGRYDDGNGRFGGGNRRFGSGGGGHDGGKG 255
R S G+ GG G G G GGG G GGGG GG G
Sbjct: 3 RQLSPFPGKTPGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGG 131
>TC88387 similar to GP|13357253|gb|AAK20050.1 putative zinc finger protein
{Oryza sativa (japonica cultivar-group)}, partial (96%)
Length = 1286
Score = 65.5 bits (158), Expect = 2e-11
Identities = 52/178 (29%), Positives = 68/178 (37%), Gaps = 11/178 (6%)
Frame = +1
Query: 105 CYTCGDTGHIARDCDRSDRNDRNDRSGGGGGG-DRDRACYTCGSFEHFARDCMRGGGNNN 163
C+ C GHIA +C G + C+TCG H AR+C
Sbjct: 481 CHNCSLPGHIASECSTKSLCWNCKEPGHMASSCPNEGICHTCGKAGHRARECTVPQKPP- 657
Query: 164 NGGGGYGGGGTSCYRCGGVGHIARDCATPSSGGGGGGACYKCGEVGHIARDCSNE----- 218
G C C GHIA +C AC C + GH+ARDC N+
Sbjct: 658 -------GDLRLCNNCYKQGHIAVECTNEK-------ACNNCRKTGHLARDCPNDPICNL 795
Query: 219 ---GGRFDGGNGRYDDGNGRFGGGNRRFGSGGGGHDGG--KGTCFNCGKAGHFARDCV 271
G + + R GGG+ R GG DGG C +C + GH +RDC+
Sbjct: 796 CNISGHVARQCPKSNVIGDRGGGGSLR----GGYRDGGFRDVVCRSCQQFGHMSRDCM 957
Score = 29.3 bits (64), Expect = 1.7
Identities = 19/71 (26%), Positives = 28/71 (38%), Gaps = 2/71 (2%)
Frame = +1
Query: 202 CYKCGEVGHIARDCSNEGGRFDGGNGRY--DDGNGRFGGGNRRFGSGGGGHDGGKGTCFN 259
C C GH R+C N + + + + + N + +G C
Sbjct: 424 CKNCKRPGHYVRECPNVAVCHNCSLPGHIASECSTKSLCWNCKEPGHMASSCPNEGICHT 603
Query: 260 CGKAGHFARDC 270
CGKAGH AR+C
Sbjct: 604 CGKAGHRAREC 636
>BQ144502 weakly similar to GP|6523547|emb hydroxyproline-rich glycoprotein
DZ-HRGP {Volvox carteri f. nagariensis}, partial (39%)
Length = 1358
Score = 64.3 bits (155), Expect = 5e-11
Identities = 74/228 (32%), Positives = 81/228 (35%), Gaps = 27/228 (11%)
Frame = +2
Query: 57 ADGDNGKTKAVDVTG-PKGEPLQVRQDNHGGGGGGRGFRGGERRNGGGGCYTCGDTGHIA 115
A G G + V V G +G PL+ GG GGGRG RGG GGG G
Sbjct: 68 AGGGGGGLRLVRVGGWGRGGPLR------GGWGGGRGPRGGGGGGPGGGGRWRGGCPRPQ 229
Query: 116 RDCDRSDRNDRND------------RSGGGGGGDRDRACYTCGSFEHFAR------DCMR 157
C R GG G G D GSF AR D R
Sbjct: 230 AAC*GEWGRGRGGGPVQGRRGAGWVGEGGAGDGALDPGVPPLGSFLTLARAPRRRPDGGR 409
Query: 158 GGGNNNNGGGGYGGGGTSCYRCGGVGHIARDCATPSS-GGGGGGACYKCGEVG----HIA 212
GG G G GGG R GG G AR GG G G GEVG +
Sbjct: 410 AGGRRGRGRWGAGGGRAGGRRGGGAGGGARPRIVEGLWGGTGAGGLAGGGEVGGGM*RLG 589
Query: 213 RDCSNEGG---RFDGGNGRYDDGNGRFGGGNRRFGSGGGGHDGGKGTC 257
R+ GG +F GR G GGG G GGG G + C
Sbjct: 590 REGRAPGGAAHQFSWQEGRVPIPGGEEGGGG---GGGGGWRRGAREGC 724
Score = 55.8 bits (133), Expect = 2e-08
Identities = 63/212 (29%), Positives = 68/212 (31%), Gaps = 34/212 (16%)
Frame = +1
Query: 85 GGGGGGRGFRGGERRNGGGGCYTCGD----TGHIARDCDRSDRNDRNDRSGGG--GGGDR 138
GG GGG G GG RR GG G G + R + GG GGG R
Sbjct: 136 GGVGGGAGAEGGRRRRARGGGAVAGGMPSAAGGLLRGVGPGAGGGPGPGAAGGRVGGGGR 315
Query: 139 DRA------CYTCGSFEHFARDCM----RGGGNNNNGGGGYGGGGTSCYRCGGVGHIARD 188
R C G F RG G G GG R G G
Sbjct: 316 GRGRGSGPRCAALGEFPDLGPGTTATP*RGARGWPAGSGALGG------RRGARGR---- 465
Query: 189 CATPSSGGGGGGACYKCGEVGHIARDCSNEGGRFDGGNGRYDDG------NGRFGG---- 238
P GGGGGGA CG +GG GG GR G G G
Sbjct: 466 --APRGGGGGGGAASDCGGA------LGGDGGGGTGGGGRGGRGYVATRTRGTSAGRGCP 621
Query: 239 ------GNRRFGSGGGGHDGG--KGTCFNCGK 262
G R GGGG GG +G C +
Sbjct: 622 PIFLARGTRSHSGGGGGGRGGGRRGVAAGCAR 717
Score = 53.1 bits (126), Expect = 1e-07
Identities = 58/183 (31%), Positives = 64/183 (34%), Gaps = 3/183 (1%)
Frame = +3
Query: 85 GGGGGGRGFRGGERRNGGGGCYTCGDTGHIARDCDRSDRNDRNDRSGGGGGGD---RDRA 141
GGGGGGR GG G GG G G GGGGGGD R R
Sbjct: 111 GGGGGGRCGGGGGGGGGRGGAEAAGPGG------------------GGGGGGDALGRRRL 236
Query: 142 CYTCGSFEHFARDCMRGGGNNNNGGGGYGGGGTSCYRCGGVGHIARDCATPSSGGGGGGA 201
G+ GGG + GGGG GG G + G + R G G
Sbjct: 237 AKGSGA--------GGGGGARSRGGGGPGGWGRAGQGTGLWTPVCRPWGVS*PWPGHHG- 389
Query: 202 CYKCGEVGHIARDCSNEGGRFDGGNGRYDDGNGRFGGGNRRFGSGGGGHDGGKGTCFNCG 261
D G R GG G G G GG R G+ GGG GG+G G
Sbjct: 390 ------------DALTGGARVAGGVG----GAGGPEGG-ARAGAAGGGRGGGRGLGLWRG 518
Query: 262 KAG 264
G
Sbjct: 519 SGG 527
Score = 51.2 bits (121), Expect = 4e-07
Identities = 67/228 (29%), Positives = 76/228 (32%), Gaps = 44/228 (19%)
Frame = +3
Query: 85 GGGGGGRGFRGGERRNG-GGGCYTCGDT---GHIARDCDRSDRNDRNDRSGGGGGGDRDR 140
GGGGGG G RGG G GGG GD +A+ R GGG GG R
Sbjct: 135 GGGGGGGGGRGGAEAAGPGGGGGGGGDALGRRRLAKGSGAGGGGGARSRGGGGPGG-WGR 311
Query: 141 ACYTCGSFEHFAR-------------DCMRGGGNNNNGGGGYGG--GGTSCYRCGGVGHI 185
A G + R D + GG G GG GG GG GG
Sbjct: 312 AGQGTGLWTPVCRPWGVS*PWPGHHGDALTGGARVAGGVGGAGGPEGGARAGAAGGGRGG 491
Query: 186 ARDCAT-PSSGGGGGGACYKCGEVGHIARDCSNEGGR----------FDGGNGRYDDGNG 234
R SGGG G ++ G A CS+ R F G + G
Sbjct: 492 GRGLGLWRGSGGGRGRGDWRGG--ARWAGVCSDSDERDERRAGLPTNFLGKRDAFPFRGG 665
Query: 235 RFGGGNRRFGSGGG--------------GHDGGKGTCFNCGKAGHFAR 268
R G G G GGG GG G C G+ AR
Sbjct: 666 RRGAGGGEEGGGGGVRARDAVRRSRAGCAWGGGAGRCGEGGRRAQHAR 809
Score = 50.4 bits (119), Expect = 7e-07
Identities = 55/181 (30%), Positives = 62/181 (33%), Gaps = 2/181 (1%)
Frame = +1
Query: 85 GGGGGGRGFRGGERRNGG--GGCYTCGDTGHIARDCDRSDRNDRNDRSGGGGGGDRDRAC 142
GGG GGRG GG R G G C G G A + R GGG
Sbjct: 37 GGGRGGRGAGGGGGRWGAQAGACGGVGAGGAAAGGVGGGAGAEGGRRRRARGGGA----- 201
Query: 143 YTCGSFEHFARDCMRGGGNNNNGGGGYGGGGTSCYRCGGVGHIARDCATPSSGGGGGGAC 202
G A +RG G GG G G G R GG G G G G C
Sbjct: 202 -VAGGMPSAAGGLLRGVGPGAGGGPGPGAAGG---RVGGGGR--------GRGRGSGPRC 345
Query: 203 YKCGEVGHIARDCSNEGGRFDGGNGRYDDGNGRFGGGNRRFGSGGGGHDGGKGTCFNCGK 262
GE + + R G G + G+G GG G G GG G +CG
Sbjct: 346 AALGEFPDLGPGTTATP*R--GARG-WPAGSGALGGRRGARGRAPRGGGGGGGAASDCGG 516
Query: 263 A 263
A
Sbjct: 517 A 519
Score = 48.9 bits (115), Expect = 2e-06
Identities = 51/146 (34%), Positives = 59/146 (39%), Gaps = 8/146 (5%)
Frame = +1
Query: 64 TKAVDVTGPKGEPLQVRQDNHGGGGGGRGFRGGERRNGGGGCYTCGDTGHIARDCDRSDR 123
T A G +G P G GG RG RG R GGGG G A DC +
Sbjct: 382 TTATP*RGARGWPA-----GSGALGGRRGARGRAPRGGGGG-------GGAASDCGGALG 525
Query: 124 NDRNDRSGGGGGGDRDR-ACYTCGSFEHFARDC-----MRGGGNNNNGGGGYGGGGTSCY 177
D +GGGG G R A T G+ R C RG +++ GGGG GGG
Sbjct: 526 GDGGGGTGGGGRGGRGYVATRTRGT--SAGRGCPPIFLARGTRSHSGGGGGGRGGG---- 687
Query: 178 RCGGVGHIARD--CATPSSGGGGGGA 201
R G AR C + G GG A
Sbjct: 688 RRGVAAGCARGMLCGARAQGAPGGVA 765
Score = 48.1 bits (113), Expect = 4e-06
Identities = 42/122 (34%), Positives = 47/122 (38%)
Frame = +3
Query: 134 GGGDRDRACYTCGSFEHFARDCMRGGGNNNNGGGGYGGGGTSCYRCGGVGHIARDCATPS 193
GGG R + G + C+ GGG GGG GGGG GG G + A P
Sbjct: 36 GGGARGAGGWRRGGEVGGSGWCVWGGGG---GGGRCGGGGG-----GGGGRGGAEAAGPG 191
Query: 194 SGGGGGGACYKCGEVGHIARDCSNEGGRFDGGNGRYDDGNGRFGGGNRRFGSGGGGHDGG 253
GGGGGG G R G+G GGG R S GGG GG
Sbjct: 192 GGGGGGGDAL---------------------GRRRLAKGSGAGGGGGAR--SRGGGGPGG 302
Query: 254 KG 255
G
Sbjct: 303 WG 308
Score = 42.0 bits (97), Expect = 3e-04
Identities = 44/147 (29%), Positives = 53/147 (35%), Gaps = 4/147 (2%)
Frame = +3
Query: 85 GGGGGGRGFRGGERRNGGGGCYTCGDTGHIARDCDRSDRNDRNDRSGG---GGGGDRDRA 141
GG GGGRG GG G A C SD ++R++R G G RD
Sbjct: 477 GGRGGGRGLGLWRGSGGGRGRGDWRGGARWAGVC--SDSDERDERRAGLPTNFLGKRDAF 650
Query: 142 CYTCGSFEHFARDCMRGGGNNNNGGGGYGGGGTSCYRCGGVGHIARDCATPSSGG-GGGG 200
+ GG GGG GGGG G ARD S G GG
Sbjct: 651 PFR--------------GGRRGAGGGEEGGGG---------GVRARDAVRRSRAGCAWGG 761
Query: 201 ACYKCGEVGHIARDCSNEGGRFDGGNG 227
+CGE G A+ + G+G
Sbjct: 762 GAGRCGEGGRRAQHARARSSGVEAGSG 842
Score = 40.8 bits (94), Expect = 6e-04
Identities = 41/122 (33%), Positives = 49/122 (39%), Gaps = 12/122 (9%)
Frame = +1
Query: 157 RGGGNNNNGGGGYGGG---GTSCYRCGGVGHIARDCATPSSGGGGGGACYKCGEVGHIAR 213
+GGG G G GGG G CGGVG ++GG GGGA + G R
Sbjct: 34 KGGGRG--GRGAGGGGGRWGAQAGACGGVG-----AGGAAAGGVGGGAGAEGGR-----R 177
Query: 214 DCSNEGGRFDGG----NGRYDDGNGRFGGGNRRFGS-----GGGGHDGGKGTCFNCGKAG 264
+ GG GG G G G GG G+ GGGG G+G+ C G
Sbjct: 178 RRARGGGAVAGGMPSAAGGLLRGVGPGAGGGPGPGAAGGRVGGGGRGRGRGSGPRCAALG 357
Query: 265 HF 266
F
Sbjct: 358 EF 363
Score = 31.2 bits (69), Expect = 0.45
Identities = 26/72 (36%), Positives = 29/72 (40%), Gaps = 5/72 (6%)
Frame = +2
Query: 191 TPSSGGGGGGACYKCGEVG--HIARDCSNEGGRFDGGNGRYDDGNGRFGGGNRRFGSGGG 248
T GG GGG G G + R GG GR G +GGG G GGG
Sbjct: 29 TRRGGGAGGGGLAAGGGGGGLRLVR---------VGGWGRGGPLRGGWGGGRGPRGGGGG 181
Query: 249 GHDGG---KGTC 257
G GG +G C
Sbjct: 182 GPGGGGRWRGGC 217
Score = 27.3 bits (59), Expect = 6.5
Identities = 51/194 (26%), Positives = 61/194 (31%), Gaps = 22/194 (11%)
Frame = +2
Query: 86 GGGGGRGFRGGERRNGGGG---------CYTCGDTGHIARDCDRSDRNDRNDRSGGGGGG 136
G GGGR GG R G GG + G +A + R R G GG
Sbjct: 440 GAGGGRA--GGRRGGGAGGGARPRIVEGLWGGTGAGGLAGGGEVGGGM*RLGREGRAPGG 613
Query: 137 DRDRACYTCGSFEHFARDCMRGGGNNNNGGGGYGGGGTSCYRCGGVGHIARDCATPSSGG 196
+ + G R + GG GGGG GGG R G +AR
Sbjct: 614 AAHQFSWQEG------RVPIPGG--EEGGGGGGGGGWRRGAREGCCAALARRVRL----- 754
Query: 197 GGGGACYKCGEVGHIARDCSNE---GGRFDGGN----------GRYDDGNGRFGGGNRRF 243
GG + G AR + E GG + G R N R R
Sbjct: 755 GGWRWAVRGGRTARAARARTIERRRGGIWGAGEVLERRRAVDLARVSARNKRVPRRQSRA 934
Query: 244 GSGGGGHDGGKGTC 257
G G G G C
Sbjct: 935 GRGAGDDGADAGRC 976
>TC86766 weakly similar to PIR|T06291|T06291 extensin homolog T9E8.80 -
Arabidopsis thaliana, partial (18%)
Length = 1260
Score = 64.3 bits (155), Expect = 5e-11
Identities = 63/183 (34%), Positives = 70/183 (37%), Gaps = 14/183 (7%)
Frame = -1
Query: 87 GGGGRGFR-----GGERRNGGGG---CYTCGDTGHIARDCDRSDRNDRNDRSGGGGGGDR 138
GGGG G+ GGE GGGG YT GD G D GGG G
Sbjct: 495 GGGGEGYAYTGDGGGEVYTGGGGEG*AYT-GDGG--------------GDGDGGG**G-- 367
Query: 139 DRACYTCGSFEHFARDCMRGGGNNNNGGGGYGGGGTSCYRCGGVGHIARDCATPSSGGGG 198
CG+ + GGG + GGGG GGG T GG G + GGGG
Sbjct: 366 ----V*CGAGDD-------GGGGSTQGGGGGGGGSTQGGGDGGGGE*TGGGGECTGGGGG 220
Query: 199 GGACYKCGEVGHIARDCSNEGGRFDGGNGRYDDGNG------RFGGGNRRFGSGGGGHDG 252
G G VG + GG GG G G G GGG G+GGGG G
Sbjct: 219 GDKYGYTGGVG----E*GGGGGEXGGGGGE*AGGGGGEL***GTGGGGENTGAGGGGGGG 52
Query: 253 GKG 255
G
Sbjct: 51 EFG 43
Score = 52.8 bits (125), Expect = 1e-07
Identities = 50/148 (33%), Positives = 57/148 (37%), Gaps = 6/148 (4%)
Frame = -1
Query: 123 RNDRNDRSGGGGGGDRDRACYTCGSFEHFARDCMRGGGNNNNGGGGYG-----GGGTSCY 177
R+ + ++ GGGGGD T G + GGG+ GGGG G GG Y
Sbjct: 612 RSSQKNQ*KGGGGGDAYEIPKTGGIGPSYTGG---GGGDEYTGGGGEGYAYTGDGGGEVY 442
Query: 178 RCGGVGHIARDCATPSSGGGGGGACYKCGEVGHIARDCSNEGGRFDGGNGRYDDGNGRFG 237
GG A G GGG CG A D DGG G G G G
Sbjct: 441 TGGGGEG*AYTGDGGGDGDGGG**GV*CG-----AGD--------DGGGGSTQGGGGGGG 301
Query: 238 GGNRRFGSGGGGH-DGGKGTCFNCGKAG 264
G + G GGGG GG G C G G
Sbjct: 300 GSTQGGGDGGGGE*TGGGGECTGGGGGG 217
Score = 46.6 bits (109), Expect = 1e-05
Identities = 47/132 (35%), Positives = 52/132 (38%), Gaps = 7/132 (5%)
Frame = -1
Query: 85 GGGGGGRGFRGGERRNGGGGCYTCGDTGHIARDCDRSDRNDRNDRSGGGGGGDRDRACYT 144
GGGGGG G G +GGGG T G + +GGGGGG D+ YT
Sbjct: 321 GGGGGGGGSTQG-GGDGGGGE*TGGG----------------GECTGGGGGG--DKYGYT 199
Query: 145 CGSFEHFARDCMRGGGNNNNGGGGYG-------GGGTSCYRCGGVGHIARDCATPSSGGG 197
G E GGG GGG G GGG GG G GGG
Sbjct: 198 GGVGE*GGGGGEXGGGGGE*AGGGGGEL***GTGGGGENTGAGGGG------GGGEFGGG 37
Query: 198 GGGACYKCGEVG 209
GGG + G VG
Sbjct: 36 GGGDLTQYGGVG 1
>TC76705 similar to SP|Q07202|CORA_MEDSA Cold and drought-regulated protein
CORA. [Alfalfa] {Medicago sativa}, partial (80%)
Length = 854
Score = 62.8 bits (151), Expect = 1e-10
Identities = 49/140 (35%), Positives = 58/140 (41%), Gaps = 10/140 (7%)
Frame = +2
Query: 124 NDRNDRSGGG--GGGDRDRACYTCGSFEHFARDCMRGGGNNNNGGGGY--GGGGTSCYRC 179
N+ ND GG GG Y G + H GG +NGGGGY GGGG +
Sbjct: 173 NEVNDAKYGGYNHGGYNHGGGYNGGGYNHGGG--YHNGGGYHNGGGGYHNGGGGYN---- 334
Query: 180 GGVGHIARDCATPSSGGGGGGACYKCG----EVGHIARDCSNEGG--RFDGGNGRYDDGN 233
GG GH G GG GA E + D N GG F+ G Y+ G
Sbjct: 335 GGGGHGGHGGYNRGGGHGGRGAAESVAVQTEEKTNEVNDARNGGGGGSFNKGGASYNHGR 514
Query: 234 GRFGGGNRRFGSGGGGHDGG 253
G + G + GGGGH GG
Sbjct: 515 GSYHHG*GGYNHGGGGHGGG 574
Score = 54.7 bits (130), Expect = 4e-08
Identities = 38/97 (39%), Positives = 50/97 (51%), Gaps = 6/97 (6%)
Frame = +2
Query: 83 NHGGG---GGGRGFRGGERRNGGGGCYTCGDTGHIARDCDRSDRNDRNDRSGGGGGG-DR 138
N GGG GGG G GG R GG G ++ + + ++ ND R+GGGGG ++
Sbjct: 314 NGGGGYNGGGGHGGHGGYNRGGGHGGRGAAESVAVQTEEKTNEVNDA--RNGGGGGSFNK 487
Query: 139 DRACYT--CGSFEHFARDCMRGGGNNNNGGGGYGGGG 173
A Y GS+ H G G N+GGGG+GGGG
Sbjct: 488 GGASYNHGRGSYHH-------G*GGYNHGGGGHGGGG 577
Score = 38.5 bits (88), Expect = 0.003
Identities = 34/105 (32%), Positives = 40/105 (37%)
Frame = +2
Query: 169 YGGGGTSCYRCGGVGHIARDCATPSSGGGGGGACYKCGEVGHIARDCSNEGGRFDGGNGR 228
YGG Y GG H GGG G Y N GG + G G
Sbjct: 194 YGG-----YNHGGYNH----------GGGYNGGGY-------------NHGGGYHNGGGY 289
Query: 229 YDDGNGRFGGGNRRFGSGGGGHDGGKGTCFNCGKAGHFARDCVEA 273
++ G G GG +GGGGH G G +N G GH R E+
Sbjct: 290 HNGGGGYHNGGGGY--NGGGGHGGHGG--YNRG-GGHGGRGAAES 409
Score = 33.1 bits (74), Expect = 0.12
Identities = 25/78 (32%), Positives = 28/78 (35%)
Frame = +2
Query: 59 GDNGKTKAVDVTGPKGEPLQVRQDNHGGGGGGRGFRGGERRNGGGGCYTCGDTGHIARDC 118
G G+ A V E D GGGGG +GG N G G Y G G+
Sbjct: 380 GHGGRGAAESVAVQTEEKTNEVNDARNGGGGGSFNKGGASYNHGRGSYHHG*GGY----- 544
Query: 119 DRSDRNDRNDRSGGGGGG 136
N GG GGG
Sbjct: 545 --------NHGGGGHGGG 574
Score = 30.8 bits (68), Expect = 0.59
Identities = 20/52 (38%), Positives = 24/52 (45%)
Frame = +2
Query: 204 KCGEVGHIARDCSNEGGRFDGGNGRYDDGNGRFGGGNRRFGSGGGGHDGGKG 255
K EV N GG GG Y+ G GGG + +GGG H+GG G
Sbjct: 167 KTNEVNDAKYGGYNHGGYNHGGG--YNGGGYNHGGG---YHNGGGYHNGGGG 307
>TC89001 similar to GP|5726567|gb|AAD48471.1| glycine-rich RNA-binding
protein {Glycine max}, partial (90%)
Length = 488
Score = 62.4 bits (150), Expect = 2e-10
Identities = 39/96 (40%), Positives = 41/96 (42%)
Frame = +3
Query: 158 GGGNNNNGGGGYGGGGTSCYRCGGVGHIARDCATPSSGGGGGGACYKCGEVGHIARDCSN 217
GGG GGGGYGGGG G GGGG Y G
Sbjct: 297 GGGGRGGGGGGYGGGG---------------------GYGGGGGGYGGG----------- 380
Query: 218 EGGRFDGGNGRYDDGNGRFGGGNRRFGSGGGGHDGG 253
GGR DGG R G G GGG+R +G GGGG GG
Sbjct: 381 -GGRRDGGYSRSGGGGGYGGGGDRGYGGGGGGGYGG 485
Score = 45.4 bits (106), Expect = 2e-05
Identities = 49/185 (26%), Positives = 62/185 (33%)
Frame = +3
Query: 16 TKGFGFIKPDDDSEDLFVHQSDIRSEGFRSLSEGDRVEFTIADGDNGKTKAVDVTGPKGE 75
++GFGF+ F + +R D +E +G+ V+ +G
Sbjct: 171 SRGFGFVT--------FAEEKSMR----------DAIEEMNGQDIDGRNITVNEAQSRGS 296
Query: 76 PLQVRQDNHGGGGGGRGFRGGERRNGGGGCYTCGDTGHIARDCDRSDRNDRNDRSGGGGG 135
R GG GGG G+ GGGG Y GGGG
Sbjct: 297 GGGGRGGGGGGYGGGGGY------GGGGGGY-------------------------GGGG 383
Query: 136 GDRDRACYTCGSFEHFARDCMRGGGNNNNGGGGYGGGGTSCYRCGGVGHIARDCATPSSG 195
G RD GG + + GGGGYGGGG Y GG G
Sbjct: 384 GRRD------------------GGYSRSGGGGGYGGGGDRGYGGGG------------GG 473
Query: 196 GGGGG 200
G GGG
Sbjct: 474 GYGGG 488
Score = 36.2 bits (82), Expect = 0.014
Identities = 18/35 (51%), Positives = 20/35 (56%)
Frame = +3
Query: 221 RFDGGNGRYDDGNGRFGGGNRRFGSGGGGHDGGKG 255
R GG GR G G GGG +G GGGG+ GG G
Sbjct: 288 RGSGGGGRGGGGGGYGGGGG--YGGGGGGYGGGGG 386
Score = 27.7 bits (60), Expect = 5.0
Identities = 23/58 (39%), Positives = 24/58 (40%), Gaps = 9/58 (15%)
Frame = +3
Query: 213 RDCSNE--GGRFDGGN-------GRYDDGNGRFGGGNRRFGSGGGGHDGGKGTCFNCG 261
RD E G DG N R G GR GGG G GGGG GG G + G
Sbjct: 216 RDAIEEMNGQDIDGRNITVNEAQSRGSGGGGRGGGGG---GYGGGGGYGGGGGGYGGG 380
>TC87868 similar to PIR|T05112|T05112 splicing factor 9G8-like SR protein
RSZp22 [validated] - Arabidopsis thaliana, partial (91%)
Length = 860
Score = 60.5 bits (145), Expect = 7e-10
Identities = 32/74 (43%), Positives = 34/74 (45%)
Frame = +3
Query: 127 NDRSGGGGGGDRDRACYTCGSFEHFARDCMRGGGNNNNGGGGYGGGGTSCYRCGGVGHIA 186
N RSGGGGGG GGG GGGG GG CY CG GH A
Sbjct: 252 NSRSGGGGGG---------------------GGGGRGRGGGGGGGSDLKCYECGEPGHFA 368
Query: 187 RDCATPSSGGGGGG 200
R+C + GGGG G
Sbjct: 369 RECR--NRGGGGAG 404
Score = 59.3 bits (142), Expect = 2e-09
Identities = 31/79 (39%), Positives = 41/79 (51%), Gaps = 5/79 (6%)
Frame = +3
Query: 68 DVTGPKGEPLQVRQDNHGGGGGGRGFRGGERRNGGGG-----CYTCGDTGHIARDCDRSD 122
D+ G G +++ ++ GGGGG G G R GGGG CY CG+ GH AR+C
Sbjct: 210 DLDGKNGWRVELSHNSRSGGGGGGGGGGRGRGGGGGGGSDLKCYECGEPGHFAREC---- 377
Query: 123 RNDRNDRSGGGGGGDRDRA 141
+R GGG G R R+
Sbjct: 378 ----RNRGGGGAGRRRSRS 422
Score = 58.9 bits (141), Expect = 2e-09
Identities = 34/71 (47%), Positives = 39/71 (54%), Gaps = 3/71 (4%)
Frame = +3
Query: 161 NNNNGGGGYGGGGTSCYRCGGVGHIARDCATPSSGGGGGGA---CYKCGEVGHIARDCSN 217
+N+ GGG GGGG GG G GGGGGG+ CY+CGE GH AR+C N
Sbjct: 249 HNSRSGGGGGGGG------GGRGR---------GGGGGGGSDLKCYECGEPGHFARECRN 383
Query: 218 EGGRFDGGNGR 228
GG GG GR
Sbjct: 384 RGG---GGAGR 407
Score = 49.7 bits (117), Expect = 1e-06
Identities = 24/50 (48%), Positives = 27/50 (54%)
Frame = +3
Query: 226 NGRYDDGNGRFGGGNRRFGSGGGGHDGGKGTCFNCGKAGHFARDCVEASG 275
N R G G GGG R G GGGG D C+ CG+ GHFAR+C G
Sbjct: 252 NSRSGGGGGGGGGGRGRGGGGGGGSDL---KCYECGEPGHFARECRNRGG 392
>AW690548 similar to SP|Q09134|GRPA Abscisic acid and environmental stress
inducible protein. [Sickle medic] {Medicago falcata},
partial (68%)
Length = 599
Score = 58.2 bits (139), Expect = 3e-09
Identities = 41/137 (29%), Positives = 56/137 (39%)
Frame = +1
Query: 124 NDRNDRSGGGGGGDRDRACYTCGSFEHFARDCMRGGGNNNNGGGGYGGGGTSCYRCGGVG 183
N+ ND GGGG Y G ++ GGG+++ GGG + GGG Y C G
Sbjct: 151 NEVNDAKYGGGGN------YHNGGGHYY------GGGSHHGGGGSHHGGGGCHYYCHG-- 288
Query: 184 HIARDCATPSSGGGGGGACYKCGEVGHIARDCSNEGGRFDGGNGRYDDGNGRFGGGNRRF 243
C + + + E + D GG + G G Y +G G + GG
Sbjct: 289 ----HCCSYAE-----FVAVQTEEKTNEVNDAKYGGGGYHNGGGNYHNGGGHYYGGGSH- 438
Query: 244 GSGGGGHDGGKGTCFNC 260
GGG H GG G + C
Sbjct: 439 -GGGGSHHGGGGCHYYC 486
Score = 51.2 bits (121), Expect = 4e-07
Identities = 35/108 (32%), Positives = 47/108 (43%), Gaps = 9/108 (8%)
Frame = +1
Query: 83 NHGGG---GGGRGFRGGERRNGGGGCY------TCGDTGHIARDCDRSDRNDRNDRSGGG 133
++GGG GGG GG +GGGGC+ C +A + N+ ND GG
Sbjct: 193 HNGGGHYYGGGSHHGGGGSHHGGGGCHYYCHGHCCSYAEFVAVQTEEKT-NEVNDAKYGG 369
Query: 134 GGGDRDRACYTCGSFEHFARDCMRGGGNNNNGGGGYGGGGTSCYRCGG 181
GG Y G ++ GG ++ GGG + GGG Y C G
Sbjct: 370 GGYHNGGGNYHNGGGHYY-------GGGSHGGGGSHHGGGGCHYYCHG 492
Score = 45.4 bits (106), Expect = 2e-05
Identities = 40/130 (30%), Positives = 44/130 (33%), Gaps = 11/130 (8%)
Frame = +1
Query: 87 GGGGRGFRGGERRNGGGGCYTCGDTGHIARDCDRSDRNDRNDRSGGGGGGDRDRACYTCG 146
GGGG GG GGG + G + H GGGG C
Sbjct: 175 GGGGNYHNGGGHYYGGGSHHGGGGSHH-------------------GGGGCHYYCHGHCC 297
Query: 147 SFEHF-----------ARDCMRGGGNNNNGGGGYGGGGTSCYRCGGVGHIARDCATPSSG 195
S+ F D GGG +NGGG Y GG Y GG H S
Sbjct: 298 SYAEFVAVQTEEKTNEVNDAKYGGGGYHNGGGNYHNGGGHYY--GGGSH-----GGGGSH 456
Query: 196 GGGGGACYKC 205
GGGG Y C
Sbjct: 457 HGGGGCHYYC 486
>TC76561 similar to PIR|S71779|S71779 glycine-rich RNA-binding protein GRP1
- wheat, partial (95%)
Length = 1054
Score = 54.3 bits (129), Expect = 5e-08
Identities = 36/95 (37%), Positives = 38/95 (39%), Gaps = 2/95 (2%)
Frame = +1
Query: 158 GGGNNNNGGGGYGGGGTSCYRCGGVGHIARDCATPSSGGGGGGACYKCGEVGHIARDCSN 217
GGG GGGGYGGGG GG G R GGGGG Y
Sbjct: 604 GGGGGGRGGGGYGGGG------GGYGGERRGYGGGGGYGGGGGGGY-------------- 723
Query: 218 EGGRFDGGNGRYD--DGNGRFGGGNRRFGSGGGGH 250
G GG G Y G G +GGG G G GG+
Sbjct: 724 --GERRGGGGGYSRGGGGGGYGGGGYSRGGGDGGY 822
Score = 54.3 bits (129), Expect = 5e-08
Identities = 47/136 (34%), Positives = 55/136 (39%), Gaps = 4/136 (2%)
Frame = +1
Query: 52 VEFTIADGDNGKTKAVDVTGPKGEPLQVRQ-DNHGGGGGGRGFRGGERRNGGGGCYTCGD 110
V F N +A++ G + V Q + G GGGG G RGG GGGG Y
Sbjct: 493 VTFANEKSMNDAIEAMNGQDLDGRNITVNQAQSRGSGGGGGGGRGGGGYGGGGGGY---- 660
Query: 111 TGHIARDCDRSDRNDRNDRSGGGGGGDRDRACYTCGSFEHFARDCMRGGG---NNNNGGG 167
+R GGGGGG +R RGGG + GGG
Sbjct: 661 ------GGERRGYGGGGGYGGGGGGGYGER----------------RGGGGGYSRGGGGG 774
Query: 168 GYGGGGTSCYRCGGVG 183
GYGGGG S R GG G
Sbjct: 775 GYGGGGYS--RGGGDG 816
Score = 45.8 bits (107), Expect = 2e-05
Identities = 34/93 (36%), Positives = 37/93 (39%)
Frame = +1
Query: 161 NNNNGGGGYGGGGTSCYRCGGVGHIARDCATPSSGGGGGGACYKCGEVGHIARDCSNEGG 220
+ +GGGG GG G Y GGGGGG G R GG
Sbjct: 589 SRGSGGGGGGGRGGGGY-----------------GGGGGG-------YGGERRGYGGGGG 696
Query: 221 RFDGGNGRYDDGNGRFGGGNRRFGSGGGGHDGG 253
GG G Y G R GGG G GGGG+ GG
Sbjct: 697 YGGGGGGGY--GERRGGGGGYSRGGGGGGYGGG 789
Score = 43.5 bits (101), Expect = 9e-05
Identities = 31/71 (43%), Positives = 33/71 (45%)
Frame = +1
Query: 130 SGGGGGGDRDRACYTCGSFEHFARDCMRGGGNNNNGGGGYGGGGTSCYRCGGVGHIARDC 189
SGGGGGG R Y G + GGG G GG GGGG R GG G+
Sbjct: 598 SGGGGGGGRGGGGYGGGGGGYGGERRGYGGGG---GYGGGGGGGYGERRGGGGGY----- 753
Query: 190 ATPSSGGGGGG 200
S GGGGGG
Sbjct: 754 ---SRGGGGGG 777
Score = 38.9 bits (89), Expect = 0.002
Identities = 28/77 (36%), Positives = 32/77 (41%), Gaps = 1/77 (1%)
Frame = +1
Query: 193 SSGGGGGGACYKCGEVGHIARDCSNEGGRFDGGNGRYDDGNGRFGGGNRRFGSGGG-GHD 251
S G GGGG GGR G G Y G G +GG R +G GGG G
Sbjct: 589 SRGSGGGGG-----------------GGR---GGGGYGGGGGGYGGERRGYGGGGGYGGG 708
Query: 252 GGKGTCFNCGKAGHFAR 268
GG G G G ++R
Sbjct: 709 GGGGYGERRGGGGGYSR 759
Score = 28.5 bits (62), Expect = 2.9
Identities = 14/24 (58%), Positives = 15/24 (62%)
Frame = +1
Query: 83 NHGGGGGGRGFRGGERRNGGGGCY 106
+ GGGGGG G GG R GG G Y
Sbjct: 754 SRGGGGGGYG-GGGYSRGGGDGGY 822
>TC76556 similar to PIR|S71779|S71779 glycine-rich RNA-binding protein GRP1
- wheat, partial (95%)
Length = 754
Score = 54.3 bits (129), Expect = 5e-08
Identities = 36/95 (37%), Positives = 38/95 (39%), Gaps = 2/95 (2%)
Frame = +2
Query: 158 GGGNNNNGGGGYGGGGTSCYRCGGVGHIARDCATPSSGGGGGGACYKCGEVGHIARDCSN 217
GGG GGGGYGGGG GG G R GGGGG Y
Sbjct: 350 GGGGGGRGGGGYGGGG------GGYGGERRGYGGGGGYGGGGGGGY-------------- 469
Query: 218 EGGRFDGGNGRYD--DGNGRFGGGNRRFGSGGGGH 250
G GG G Y G G +GGG G G GG+
Sbjct: 470 --GERRGGGGGYSRGGGGGGYGGGGYSRGGGDGGY 568
Score = 54.3 bits (129), Expect = 5e-08
Identities = 47/136 (34%), Positives = 55/136 (39%), Gaps = 4/136 (2%)
Frame = +2
Query: 52 VEFTIADGDNGKTKAVDVTGPKGEPLQVRQ-DNHGGGGGGRGFRGGERRNGGGGCYTCGD 110
V F N +A++ G + V Q + G GGGG G RGG GGGG Y
Sbjct: 239 VTFANEKSMNDAIEAMNGQDLDGRNITVNQAQSRGSGGGGGGGRGGGGYGGGGGGY---- 406
Query: 111 TGHIARDCDRSDRNDRNDRSGGGGGGDRDRACYTCGSFEHFARDCMRGGG---NNNNGGG 167
+R GGGGGG +R RGGG + GGG
Sbjct: 407 ------GGERRGYGGGGGYGGGGGGGYGER----------------RGGGGGYSRGGGGG 520
Query: 168 GYGGGGTSCYRCGGVG 183
GYGGGG S R GG G
Sbjct: 521 GYGGGGYS--RGGGDG 562
Score = 45.8 bits (107), Expect = 2e-05
Identities = 34/93 (36%), Positives = 37/93 (39%)
Frame = +2
Query: 161 NNNNGGGGYGGGGTSCYRCGGVGHIARDCATPSSGGGGGGACYKCGEVGHIARDCSNEGG 220
+ +GGGG GG G Y GGGGGG G R GG
Sbjct: 335 SRGSGGGGGGGRGGGGY-----------------GGGGGG-------YGGERRGYGGGGG 442
Query: 221 RFDGGNGRYDDGNGRFGGGNRRFGSGGGGHDGG 253
GG G Y G R GGG G GGGG+ GG
Sbjct: 443 YGGGGGGGY--GERRGGGGGYSRGGGGGGYGGG 535
Score = 43.5 bits (101), Expect = 9e-05
Identities = 31/71 (43%), Positives = 33/71 (45%)
Frame = +2
Query: 130 SGGGGGGDRDRACYTCGSFEHFARDCMRGGGNNNNGGGGYGGGGTSCYRCGGVGHIARDC 189
SGGGGGG R Y G + GGG G GG GGGG R GG G+
Sbjct: 344 SGGGGGGGRGGGGYGGGGGGYGGERRGYGGGG---GYGGGGGGGYGERRGGGGGY----- 499
Query: 190 ATPSSGGGGGG 200
S GGGGGG
Sbjct: 500 ---SRGGGGGG 523
Score = 38.9 bits (89), Expect = 0.002
Identities = 28/77 (36%), Positives = 32/77 (41%), Gaps = 1/77 (1%)
Frame = +2
Query: 193 SSGGGGGGACYKCGEVGHIARDCSNEGGRFDGGNGRYDDGNGRFGGGNRRFGSGGG-GHD 251
S G GGGG GGR G G Y G G +GG R +G GGG G
Sbjct: 335 SRGSGGGGG-----------------GGR---GGGGYGGGGGGYGGERRGYGGGGGYGGG 454
Query: 252 GGKGTCFNCGKAGHFAR 268
GG G G G ++R
Sbjct: 455 GGGGYGERRGGGGGYSR 505
Score = 28.5 bits (62), Expect = 2.9
Identities = 14/24 (58%), Positives = 15/24 (62%)
Frame = +2
Query: 83 NHGGGGGGRGFRGGERRNGGGGCY 106
+ GGGGGG G GG R GG G Y
Sbjct: 500 SRGGGGGGYG-GGGYSRGGGDGGY 568
>TC76557 similar to PIR|S71779|S71779 glycine-rich RNA-binding protein GRP1
- wheat, partial (95%)
Length = 912
Score = 54.3 bits (129), Expect = 5e-08
Identities = 36/95 (37%), Positives = 38/95 (39%), Gaps = 2/95 (2%)
Frame = +1
Query: 158 GGGNNNNGGGGYGGGGTSCYRCGGVGHIARDCATPSSGGGGGGACYKCGEVGHIARDCSN 217
GGG GGGGYGGGG GG G R GGGGG Y
Sbjct: 508 GGGGGGRGGGGYGGGG------GGYGGERRGYGGGGGYGGGGGGGY-------------- 627
Query: 218 EGGRFDGGNGRYD--DGNGRFGGGNRRFGSGGGGH 250
G GG G Y G G +GGG G G GG+
Sbjct: 628 --GERRGGGGGYSRGGGGGGYGGGGYSRGGGDGGY 726
Score = 54.3 bits (129), Expect = 5e-08
Identities = 47/136 (34%), Positives = 55/136 (39%), Gaps = 4/136 (2%)
Frame = +1
Query: 52 VEFTIADGDNGKTKAVDVTGPKGEPLQVRQ-DNHGGGGGGRGFRGGERRNGGGGCYTCGD 110
V F N +A++ G + V Q + G GGGG G RGG GGGG Y
Sbjct: 397 VTFANEKSMNDAIEAMNGQDLDGRNITVNQAQSRGSGGGGGGGRGGGGYGGGGGGY---- 564
Query: 111 TGHIARDCDRSDRNDRNDRSGGGGGGDRDRACYTCGSFEHFARDCMRGGG---NNNNGGG 167
+R GGGGGG +R RGGG + GGG
Sbjct: 565 ------GGERRGYGGGGGYGGGGGGGYGER----------------RGGGGGYSRGGGGG 678
Query: 168 GYGGGGTSCYRCGGVG 183
GYGGGG S R GG G
Sbjct: 679 GYGGGGYS--RGGGDG 720
Score = 45.8 bits (107), Expect = 2e-05
Identities = 34/93 (36%), Positives = 37/93 (39%)
Frame = +1
Query: 161 NNNNGGGGYGGGGTSCYRCGGVGHIARDCATPSSGGGGGGACYKCGEVGHIARDCSNEGG 220
+ +GGGG GG G Y GGGGGG G R GG
Sbjct: 493 SRGSGGGGGGGRGGGGY-----------------GGGGGG-------YGGERRGYGGGGG 600
Query: 221 RFDGGNGRYDDGNGRFGGGNRRFGSGGGGHDGG 253
GG G Y G R GGG G GGGG+ GG
Sbjct: 601 YGGGGGGGY--GERRGGGGGYSRGGGGGGYGGG 693
Score = 43.5 bits (101), Expect = 9e-05
Identities = 31/71 (43%), Positives = 33/71 (45%)
Frame = +1
Query: 130 SGGGGGGDRDRACYTCGSFEHFARDCMRGGGNNNNGGGGYGGGGTSCYRCGGVGHIARDC 189
SGGGGGG R Y G + GGG G GG GGGG R GG G+
Sbjct: 502 SGGGGGGGRGGGGYGGGGGGYGGERRGYGGGG---GYGGGGGGGYGERRGGGGGY----- 657
Query: 190 ATPSSGGGGGG 200
S GGGGGG
Sbjct: 658 ---SRGGGGGG 681
Score = 38.9 bits (89), Expect = 0.002
Identities = 28/77 (36%), Positives = 32/77 (41%), Gaps = 1/77 (1%)
Frame = +1
Query: 193 SSGGGGGGACYKCGEVGHIARDCSNEGGRFDGGNGRYDDGNGRFGGGNRRFGSGGG-GHD 251
S G GGGG GGR G G Y G G +GG R +G GGG G
Sbjct: 493 SRGSGGGGG-----------------GGR---GGGGYGGGGGGYGGERRGYGGGGGYGGG 612
Query: 252 GGKGTCFNCGKAGHFAR 268
GG G G G ++R
Sbjct: 613 GGGGYGERRGGGGGYSR 663
Score = 28.5 bits (62), Expect = 2.9
Identities = 14/24 (58%), Positives = 15/24 (62%)
Frame = +1
Query: 83 NHGGGGGGRGFRGGERRNGGGGCY 106
+ GGGGGG G GG R GG G Y
Sbjct: 658 SRGGGGGGYG-GGGYSRGGGDGGY 726
>TC76704 homologue to SP|Q09134|GRPA_MEDFA Abscisic acid and environmental
stress inducible protein. [Sickle medic] {Medicago
falcata}, partial (81%)
Length = 771
Score = 53.5 bits (127), Expect = 8e-08
Identities = 42/114 (36%), Positives = 51/114 (43%), Gaps = 7/114 (6%)
Frame = +2
Query: 154 DCMRGGGNNNNG---GGGYGGGGTSCYRCGGVGHIARDCATPSSGGGGGGACYKCGEVGH 210
D GGG N G GGGY GGG Y GG G+ ++GGG Y G G+
Sbjct: 179 DAKYGGGYNGGGYNHGGGYNGGG---YNHGGGGY--------NNGGG-----YNHGGGGY 310
Query: 211 IARDCSNEGGRFDGG----NGRYDDGNGRFGGGNRRFGSGGGGHDGGKGTCFNC 260
+N GG GG G Y+ G G + GG G GG H GG G ++C
Sbjct: 311 -----NNGGGYNHGGGGYNGGGYNHGGGGYNGGGYNHGGGGYNHGGG-GCQYHC 454
Score = 52.4 bits (124), Expect = 2e-07
Identities = 42/128 (32%), Positives = 52/128 (39%)
Frame = +2
Query: 78 QVRQDNHGGGGGGRGFRGGERRNGGGGCYTCGDTGHIARDCDRSDRNDRNDRSGGGGGGD 137
+V +GGG G G+ G NGGG Y G G+ N+ + GGGG +
Sbjct: 170 EVNDAKYGGGYNGGGYNHGGGYNGGG--YNHGGGGY----------NNGGGYNHGGGGYN 313
Query: 138 RDRACYTCGSFEHFARDCMRGGGNNNNGGGGYGGGGTSCYRCGGVGHIARDCATPSSGGG 197
G + H GG N+GGGGY GGG Y GG G+ G
Sbjct: 314 NG------GGYNHGGGG--YNGGGYNHGGGGYNGGG---YNHGGGGY----------NHG 430
Query: 198 GGGACYKC 205
GGG Y C
Sbjct: 431 GGGCQYHC 454
Score = 49.7 bits (117), Expect = 1e-06
Identities = 40/127 (31%), Positives = 49/127 (38%), Gaps = 3/127 (2%)
Frame = +2
Query: 124 NDRNDRSGGGG---GGDRDRACYTCGSFEHFARDCMRGGGNNNNGGGGYGGGGTSCYRCG 180
N+ ND GGG GG Y G + H GGG NN GG +GGGG + G
Sbjct: 167 NEVNDAKYGGGYNGGGYNHGGGYNGGGYNH------GGGGYNNGGGYNHGGGGYN--NGG 322
Query: 181 GVGHIARDCATPSSGGGGGGACYKCGEVGHIARDCSNEGGRFDGGNGRYDDGNGRFGGGN 240
G H GGG G Y G GG ++GG + G GGG
Sbjct: 323 GYNH---------GGGGYNGGGYNHG------------GGGYNGGGYNHGGGGYNHGGGG 439
Query: 241 RRFGSGG 247
++ G
Sbjct: 440 CQYHCHG 460
>TC87796 weakly similar to SP|P03211|EBN1_EBV EBNA-1 nuclear protein.
[strain B95-8 Human herpesvirus 4] {Epstein-barr
virus}, partial (23%)
Length = 431
Score = 53.5 bits (127), Expect = 8e-08
Identities = 36/99 (36%), Positives = 39/99 (39%)
Frame = +1
Query: 166 GGGYGGGGTSCYRCGGVGHIARDCATPSSGGGGGGACYKCGEVGHIARDCSNEGGRFDGG 225
GGGYGGGG SGGGGGG VG+ GG GG
Sbjct: 13 GGGYGGGG-------------------GSGGGGGGGAGGAHGVGY------GSGGGTGGG 117
Query: 226 NGRYDDGNGRFGGGNRRFGSGGGGHDGGKGTCFNCGKAG 264
G G G GGG+ G GGG H G G G+ G
Sbjct: 118 YGGGSPGGGGGGGGSGGGGGGGGAHGGAYGGGIGGGEGG 234
Score = 48.9 bits (115), Expect = 2e-06
Identities = 39/99 (39%), Positives = 43/99 (43%), Gaps = 1/99 (1%)
Frame = +1
Query: 158 GGGNNNNGGGGYGGGGTSCYRCGGVGHIARDCATPSSGGGGGGACYKCGEVGHIARDCSN 217
GGG + GGGG G GG GVG+ SGGG GG Y G G
Sbjct: 25 GGGGGSGGGGGGGAGGAH-----GVGY--------GSGGGTGGG-YGGGSPG-------- 138
Query: 218 EGGRFDGGNGRYDDGNGRFGGGNRRFGSG-GGGHDGGKG 255
GG GG+G G G GG +G G GGG GG G
Sbjct: 139 -GGGGGGGSGGGGGGGGAHGGA---YGGGIGGGEGGGHG 243
Score = 45.1 bits (105), Expect = 3e-05
Identities = 41/118 (34%), Positives = 45/118 (37%)
Frame = +1
Query: 83 NHGGGGGGRGFRGGERRNGGGGCYTCGDTGHIARDCDRSDRNDRNDRSGGGGGGDRDRAC 142
+HGGG GG G GG GGGG G+ SGGG GG
Sbjct: 7 DHGGGYGGGGGSGG---GGGGGAGGAHGVGY---------------GSGGGTGGG----- 117
Query: 143 YTCGSFEHFARDCMRGGGNNNNGGGGYGGGGTSCYRCGGVGHIARDCATPSSGGGGGG 200
Y GS GGG +GGGG GGG GG+G GGG GG
Sbjct: 118 YGGGS-------PGGGGGGGGSGGGGGGGGAHGGAYGGGIG--------GGEGGGHGG 246
Score = 30.0 bits (66), Expect = 1.0
Identities = 13/31 (41%), Positives = 15/31 (47%)
Frame = +1
Query: 231 DGNGRFGGGNRRFGSGGGGHDGGKGTCFNCG 261
D G +GGG G GGGG G G + G
Sbjct: 7 DHGGGYGGGGGSGGGGGGGAGGAHGVGYGSG 99
Score = 27.7 bits (60), Expect = 5.0
Identities = 14/29 (48%), Positives = 15/29 (51%)
Frame = +1
Query: 85 GGGGGGRGFRGGERRNGGGGCYTCGDTGH 113
GGGGGG G GG G GG G G+
Sbjct: 163 GGGGGGGGAHGGAYGGGIGGGEGGGHGGY 249
>TC87201 similar to PIR|F86388|F86388 hypothetical protein AAG29221.1
[imported] - Arabidopsis thaliana, partial (41%)
Length = 991
Score = 53.1 bits (126), Expect = 1e-07
Identities = 64/191 (33%), Positives = 71/191 (36%), Gaps = 20/191 (10%)
Frame = -3
Query: 85 GGGGGGRGFRGGERRNGGGGC----YTCGDTGHIARDCDRSDRNDRNDRSGGGGGGDRDR 140
G GGG GG+ GGG Y G G + + + GGGGGD
Sbjct: 599 GAGGGDL*IYGGDS*TYGGGIGL**YGGGGDG---------EGGGGDS*T*GGGGGDL*T 447
Query: 141 ACYTCGSFEHFARDCMRGGGNNN---NGGGG----YGGGGTSCYRCGGVGHIARDCATPS 193
C GGG ++ GGGG YGGGG Y GG G
Sbjct: 446 YC---------------GGGGDS*TYGGGGGDL*TYGGGGGDLYTYGGGGD--------G 336
Query: 194 SGGGG-----GGACYKCGEVGHIARDCSNEGGRFDGGNGRYDDGNGRFGGGNRRFGSGGG 248
GGGG GG Y G G D GG DGG G DG GGG + GGG
Sbjct: 335 DGGGGDL*TYGGGVYTYGGGGG---DL*IYGGGGDGGGG---DG*T*GGGGGDLYTYGGG 174
Query: 249 GHD----GGKG 255
G D GG G
Sbjct: 173 GGDL*IYGGGG 141
Score = 43.9 bits (102), Expect = 7e-05
Identities = 46/130 (35%), Positives = 52/130 (39%), Gaps = 13/130 (10%)
Frame = -3
Query: 84 HGGGGGGRGFRGGERRNGGGGCYTC-GDTGHIARDCDRSDRNDRNDRSGGGGGGDRDRAC 142
+GGGG G G GG+ GGG YT G G + D + + GGGGGD
Sbjct: 356 YGGGGDGDG-GGGDL*TYGGGVYTYGGGGGDL*IYGGGGDGGGGDG*T*GGGGGD----L 192
Query: 143 YTCGSFEHFARDCMRGGGNN--NNGGGGYGGGGTS----------CYRCGGVGHIARDCA 190
YT G GGG + GGGG GGGG C R GG G
Sbjct: 191 YTYG-----------GGGGDL*IYGGGGDGGGGDG*T*GGARVVICIRIGGGG------- 66
Query: 191 TPSSGGGGGG 200
GGGG G
Sbjct: 65 ---DGGGGDG 45
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.317 0.145 0.476
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 9,445,713
Number of Sequences: 36976
Number of extensions: 180052
Number of successful extensions: 7822
Number of sequences better than 10.0: 378
Number of HSP's better than 10.0 without gapping: 1880
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 3992
length of query: 275
length of database: 9,014,727
effective HSP length: 95
effective length of query: 180
effective length of database: 5,502,007
effective search space: 990361260
effective search space used: 990361260
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (22.0 bits)
S2: 57 (26.6 bits)
Medicago: description of AC121233.14


