

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
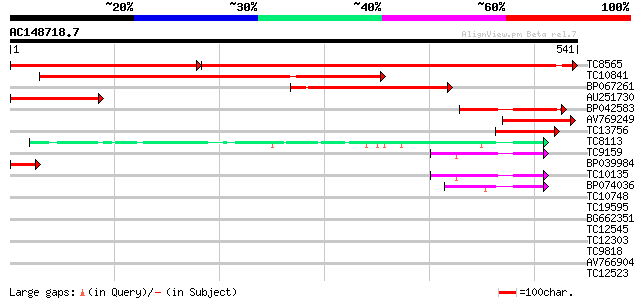
Query= AC148718.7 + phase: 0
(541 letters)
Database: LJGI
28,460 sequences; 14,692,800 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC8565 homologue to UP|Q9AVQ9 (Q9AVQ9) Phosphate transporter, pa... 563 0.0
TC10841 homologue to UP|Q9AU01 (Q9AU01) Phosphate transporter 1,... 516 e-147
BP067261 237 4e-63
AU251730 160 6e-40
BP042583 122 1e-28
AV769249 93 1e-19
TC13756 similar to UP|Q9ARI9 (Q9ARI9) Phosphate transporter 1, p... 92 3e-19
TC8113 weakly similar to UP|O23213 (O23213) Sugar transporter li... 58 3e-09
TC9159 similar to UP|HXT4_YEAST (P32467) Low-affinity glucose tr... 52 3e-07
BP039984 48 5e-06
TC10135 45 4e-05
BP074036 40 7e-04
TC10748 similar to UP|Q7XA50 (Q7XA50) Sorbitol-like transporter,... 40 0.001
TC19595 38 0.005
BG662351 38 0.005
TC12545 similar to UP|Q7XA50 (Q7XA50) Sorbitol-like transporter,... 35 0.024
TC12303 similar to GB|CAB10424.1|2245004|ATFCA6 membrane transpo... 34 0.069
TC9818 similar to UP|Q7XA50 (Q7XA50) Sorbitol-like transporter, ... 34 0.069
AV766904 32 0.20
TC12523 31 0.44
>TC8565 homologue to UP|Q9AVQ9 (Q9AVQ9) Phosphate transporter, partial
(97%)
Length = 1913
Score = 563 bits (1452), Expect(2) = 0.0
Identities = 271/359 (75%), Positives = 313/359 (86%), Gaps = 1/359 (0%)
Frame = +2
Query: 184 LVVSTAFDHAFKAPPYKVDAAASLV-PEADYVWRIILMFGAVPAGLTYYWRMKMPETARY 242
L+VS +FDH + P Y+ D AAS+V P DYVWRIILMFGA+PA LTYYWRMKMPETARY
Sbjct: 641 LIVSASFDHKYNVPSYQEDPAASMVLPAFDYVWRIILMFGALPAALTYYWRMKMPETARY 820
Query: 243 TALVAKNAKQAAQDMSSVLQVEIEAEQEKVDKIGVQDKNSFGLFSKEFLRRHGLHLLGTT 302
TALVAKNAKQAA DMS VLQVE+EAE+EKV+KI + FGLFSK+F RHG+HLLGTT
Sbjct: 821 TALVAKNAKQAASDMSKVLQVELEAEEEKVEKILESENQQFGLFSKQFASRHGMHLLGTT 1000
Query: 303 STWFLLDIAYYSSNLFQKDIYSSIGWLPPAQDMNAIHEVFKVARATTLIALCGTVPGYWF 362
+TWFLLDIA+YS NLFQKDI+S+IGW+PPA++MNAIHEV+K+ARA TLIALC TVPGYWF
Sbjct: 1001TTWFLLDIAFYSQNLFQKDIFSAIGWIPPAKEMNAIHEVYKIARAQTLIALCSTVPGYWF 1180
Query: 363 TVAFIDVIGRFAIQLMGFFFMTVFMFALAIPYDHWTKRENRIGFLVMYAMTFFFANFGPN 422
TVAFID +GRFAIQLMGFFFMTVFMFALA+PYDHWTK++NR GF+ MYA+TFFFANFGPN
Sbjct: 1181TVAFIDYMGRFAIQLMGFFFMTVFMFALALPYDHWTKKDNRFGFVAMYALTFFFANFGPN 1360
Query: 423 STTFVVPAEIFPARLRSTCHGISSAAGKAGAIIGAFGFLYASQSKDPKKRDAGYPAGIGM 482
+TTFVVPAEIFPARLRSTCHGIS+AAGKAGAI+GAFGFLYASQSKDP K D GYP GIG+
Sbjct: 1361ATTFVVPAEIFPARLRSTCHGISAAAGKAGAIVGAFGFLYASQSKDPAKTDPGYPTGIGV 1540
Query: 483 KNTLILLAVVNCLGIFFTFLVPEANGKSLEEMSGENEEEEETEAAVVDPQASSNRTVPV 541
+N+LI+L V+N LG+ FTFLVPE+ GKSLEE+SGE EE+ V +A+ +RTVPV
Sbjct: 1541RNSLIMLGVINFLGMVFTFLVPESKGKSLEELSGETEED-----GVEAIEAAGSRTVPV 1702
Score = 305 bits (781), Expect(2) = 0.0
Identities = 151/184 (82%), Positives = 161/184 (87%), Gaps = 1/184 (0%)
Frame = +3
Query: 1 MARE-QLGVLTALDAAKTQWYHFTAIIIAGMGFFTDAYDLFCIPNVTKLLGRIYYTHPGA 59
MARE +GVL ALD AKTQWYHFTAI+IAGMGFFTDAYDLFCI VTKLLGRIYYT
Sbjct: 90 MAREGTIGVLNALDVAKTQWYHFTAIVIAGMGFFTDAYDLFCISLVTKLLGRIYYTDITK 269
Query: 60 LKPGTLPPNVSAAVNGVALCGTLAGQLFFGWLGDKMGRKKVYGLTLALMVFSSLASGLSF 119
+PG LP NV AAV GVALCGTLAGQLFFGWLGDKMGRK+VYGLTL +MV S+ASGLSF
Sbjct: 270 SRPGVLPANVQAAVTGVALCGTLAGQLFFGWLGDKMGRKRVYGLTLMIMVVCSIASGLSF 449
Query: 120 GHSAKGTIGTLCFFRFWLGFGIGGDYPLSATIMSEYANKKTRGAFIASVFAMQGFGILAG 179
G+S K + TLCFFRFWLGFGIGGDYPLSATIMSEYANKKTRGAFIA+VFAMQGFGIL G
Sbjct: 450 GNSPKSVMATLCFFRFWLGFGIGGDYPLSATIMSEYANKKTRGAFIAAVFAMQGFGILGG 629
Query: 180 GIVS 183
GIV+
Sbjct: 630 GIVA 641
Score = 27.7 bits (60), Expect = 4.9
Identities = 30/133 (22%), Positives = 45/133 (33%), Gaps = 2/133 (1%)
Frame = +2
Query: 69 VSAAVNGVALCGTLAGQLFFGWLGDKMGRKKVYGLTLALMVFSSLASGLSFGHSAK--GT 126
++ A +ALC T+ G F D MGR + + M A L + H K
Sbjct: 1124 IARAQTLIALCSTVPGYWFTVAFIDYMGRFAIQLMGFFFMTVFMFALALPYDHWTKKDNR 1303
Query: 127 IGTLCFFRFWLGFGIGGDYPLSATIMSEYANKKTRGAFIASVFAMQGFGILAGGIVSLVV 186
G + + F G + + +E + R A G + G L
Sbjct: 1304 FGFVAMYALTFFFANFGPNATTFVVPAEIFPARLRSTCHGISAAAGKAGAIVGAFGFLYA 1483
Query: 187 STAFDHAFKAPPY 199
S + D A P Y
Sbjct: 1484 SQSKDPAKTDPGY 1522
>TC10841 homologue to UP|Q9AU01 (Q9AU01) Phosphate transporter 1, partial
(59%)
Length = 991
Score = 516 bits (1328), Expect = e-147
Identities = 249/330 (75%), Positives = 280/330 (84%)
Frame = +3
Query: 29 GMGFFTDAYDLFCIPNVTKLLGRIYYTHPGALKPGTLPPNVSAAVNGVALCGTLAGQLFF 88
GMGFFTDAYDLFCI VTKLLGRIYY GA KPGTLPPNVSAAVNGVA CGTL+GQLFF
Sbjct: 3 GMGFFTDAYDLFCISLVTKLLGRIYYHVDGAAKPGTLPPNVSAAVNGVAFCGTLSGQLFF 182
Query: 89 GWLGDKMGRKKVYGLTLALMVFSSLASGLSFGHSAKGTIGTLCFFRFWLGFGIGGDYPLS 148
GWLGDK+GRKKVYG+TL +MV S+ SGLSFGHS K + TLCFFRFWLGFGIGGDYPLS
Sbjct: 183 GWLGDKLGRKKVYGMTLMMMVICSIGSGLSFGHSPKSVMATLCFFRFWLGFGIGGDYPLS 362
Query: 149 ATIMSEYANKKTRGAFIASVFAMQGFGILAGGIVSLVVSTAFDHAFKAPPYKVDAAASLV 208
ATIMSEY+NKKTRGAFIA+VFAMQGFGIL GGI ++++S AF F APPY+VD S V
Sbjct: 363 ATIMSEYSNKKTRGAFIAAVFAMQGFGILGGGIFAIIISAAFKAKFDAPPYEVDPVGSTV 542
Query: 209 PEADYVWRIILMFGAVPAGLTYYWRMKMPETARYTALVAKNAKQAAQDMSSVLQVEIEAE 268
P+ADY+WRII+M GA+PA LTYYWRMKMPETARYTALVAKN +QAA+DMS VLQVEI+AE
Sbjct: 543 PQADYIWRIIVMVGALPAALTYYWRMKMPETARYTALVAKNTEQAAKDMSKVLQVEIQAE 722
Query: 269 QEKVDKIGVQDKNSFGLFSKEFLRRHGLHLLGTTSTWFLLDIAYYSSNLFQKDIYSSIGW 328
K N+F LFSKEF+RRHGLHLLGT STWFLLDIA+YS NLFQKDI+S+IGW
Sbjct: 723 ----PKGDQAQANTFALFSKEFMRRHGLHLLGTASTWFLLDIAFYSQNLFQKDIFSAIGW 890
Query: 329 LPPAQDMNAIHEVFKVARATTLIALCGTVP 358
+PPA+ MNA+ EV+++ARA TLIALC P
Sbjct: 891 IPPAKTMNALEEVYRIARAQTLIALCSYCP 980
Score = 26.9 bits (58), Expect = 8.4
Identities = 19/72 (26%), Positives = 28/72 (38%)
Frame = +3
Query: 344 VARATTLIALCGTVPGYWFTVAFIDVIGRFAIQLMGFFFMTVFMFALAIPYDHWTKRENR 403
V+ A +A CGT+ G F D +GR + M M + + + H K
Sbjct: 123 VSAAVNGVAFCGTLSGQLFFGWLGDKLGRKKVYGMTLMMMVICSIGSGLSFGHSPKS--- 293
Query: 404 IGFLVMYAMTFF 415
VM + FF
Sbjct: 294 ----VMATLCFF 317
>BP067261
Length = 460
Score = 237 bits (604), Expect = 4e-63
Identities = 111/154 (72%), Positives = 131/154 (84%)
Frame = +1
Query: 269 QEKVDKIGVQDKNSFGLFSKEFLRRHGLHLLGTTSTWFLLDIAYYSSNLFQKDIYSSIGW 328
QEK+ KI D N ++S + LLGT+STWFLLDIA+YS NLFQKDI+S+IGW
Sbjct: 1 QEKLQKITEADNNKL-VYSARNCQAPWAALLGTSSTWFLLDIAFYSQNLFQKDIFSAIGW 177
Query: 329 LPPAQDMNAIHEVFKVARATTLIALCGTVPGYWFTVAFIDVIGRFAIQLMGFFFMTVFMF 388
+PPA++MNAIHEV+K+ARA TLIALC TVPGYWFTVA ID +GRFAIQLMGFFFMTVFMF
Sbjct: 178 IPPAKEMNAIHEVYKIARAQTLIALCSTVPGYWFTVALIDYMGRFAIQLMGFFFMTVFMF 357
Query: 389 ALAIPYDHWTKRENRIGFLVMYAMTFFFANFGPN 422
ALAIPYDHWT+++NRIGF+ MY++TFFFANFGPN
Sbjct: 358 ALAIPYDHWTEKDNRIGFVAMYSLTFFFANFGPN 459
>AU251730
Length = 336
Score = 160 bits (404), Expect = 6e-40
Identities = 78/89 (87%), Positives = 80/89 (89%)
Frame = +1
Query: 1 MAREQLGVLTALDAAKTQWYHFTAIIIAGMGFFTDAYDLFCIPNVTKLLGRIYYTHPGAL 60
MAR +LGVLTALD AKTQWYHFTAIII GMGFFTDAYDLF IPNVTKLLGRIYYT G+
Sbjct: 70 MARGELGVLTALDTAKTQWYHFTAIIITGMGFFTDAYDLFSIPNVTKLLGRIYYTKEGSP 249
Query: 61 KPGTLPPNVSAAVNGVALCGTLAGQLFFG 89
KPGTLP NVS AVNGVALCGTLAGQLFFG
Sbjct: 250 KPGTLPANVSVAVNGVALCGTLAGQLFFG 336
>BP042583
Length = 459
Score = 122 bits (306), Expect = 1e-28
Identities = 66/102 (64%), Positives = 75/102 (72%)
Frame = -3
Query: 430 AEIFPARLRSTCHGISSAAGKAGAIIGAFGFLYASQSKDPKKRDAGYPAGIGMKNTLILL 489
AEIFP RLRSTCHGIS+AAGKAGA++G+FGFLYA + IG++N LILL
Sbjct: 457 AEIFPPRLRSTCHGISAAAGKAGAMVGSFGFLYAQNA-------------IGLRNVLILL 317
Query: 490 AVVNCLGIFFTFLVPEANGKSLEEMSGENEEEEETEAAVVDP 531
V N LG+FFTFLVPE NGKSLEE+SGE EEEE AA DP
Sbjct: 316 GVANLLGLFFTFLVPEPNGKSLEEISGEAEEEE--TAATRDP 197
>AV769249
Length = 485
Score = 93.2 bits (230), Expect = 1e-19
Identities = 49/71 (69%), Positives = 54/71 (76%), Gaps = 1/71 (1%)
Frame = -1
Query: 471 KRDAGYPAGIGMKNTLILLAVVNCLGIFFTFLVPEANGKSLEEMSGENEEE-EETEAAVV 529
K DAGYPAGIG+KN+L+LL VVN LG F TFLVPEA GKSLEEMSGENEEE E
Sbjct: 479 KADAGYPAGIGVKNSLLLLGVVNILGFFCTFLVPEAKGKSLEEMSGENEEEGENGTKEES 300
Query: 530 DPQASSNRTVP 540
+P + S RTVP
Sbjct: 299 EPHSFSTRTVP 267
>TC13756 similar to UP|Q9ARI9 (Q9ARI9) Phosphate transporter 1, partial
(11%)
Length = 446
Score = 91.7 bits (226), Expect = 3e-19
Identities = 44/61 (72%), Positives = 51/61 (83%)
Frame = +3
Query: 464 SQSKDPKKRDAGYPAGIGMKNTLILLAVVNCLGIFFTFLVPEANGKSLEEMSGENEEEEE 523
+Q++D K DAGYPAGIG+KN+L+LL VVN LG FTFLVPEA GKSLEEMSGE EEE E
Sbjct: 6 AQNQDKSKADAGYPAGIGVKNSLLLLGVVNILGFLFTFLVPEAKGKSLEEMSGEQEEEVE 185
Query: 524 T 524
+
Sbjct: 186 S 188
>TC8113 weakly similar to UP|O23213 (O23213) Sugar transporter like
protein, partial (93%)
Length = 1865
Score = 58.2 bits (139), Expect = 3e-09
Identities = 120/517 (23%), Positives = 195/517 (37%), Gaps = 22/517 (4%)
Frame = +2
Query: 20 YHFTAIIIAGMGFFTDAYDLFCIPNVTKLLGRIYYTHPGALKPGTLPPNVSAAVNGVALC 79
Y +I+A M YD + G + + T ++ +N AL
Sbjct: 113 YALACVIVASMVSIISGYD------TGVMSGALLFIKEDIGISDTQQEVLAGILNICALV 274
Query: 80 GTLAGQLFFGWLGDKMGRKKVYGLTLALMVFSSLASGLSFGHSAKGTIGTLCFFRFWLGF 139
G LA G D +GR+ Y + LA ++F A + +G + L F R G
Sbjct: 275 GCLAA----GKTSDYIGRR--YTIFLASILFLVGAVFMGYGPN----FAILMFGRCVCGL 424
Query: 140 GIGGDYPLSATIMSEYANKKTRGAFIASVFAMQGFGILAGGIVSLVVSTAFDHAFKAPPY 199
G+G + +E ++ TRG + G GI G I + +
Sbjct: 425 GVGFALTTAPVYSAELSSASTRGFLTSLPEVCIGLGIFIGYISNYFLGKL---------- 574
Query: 200 KVDAAASLVPEADYVWRIILMFGAVPAGLTYYWRMKMPETARYTALVAKN--AKQAAQDM 257
A +L WR++L A+P+ + MPE+ R+ + + AK+ ++
Sbjct: 575 ----ALTLG------WRLMLGLAAIPSLGLALGILTMPESPRWLVMQGRLGCAKKVLLEV 724
Query: 258 SSVLQVEIEAEQEKVDKIGVQDKNSFGLFSKEFLRRHGLHLLGTTSTWFLLDIAYYSSNL 317
S+ + E E + D+ F K+ + H H G FL L
Sbjct: 725 SNTTE-EAELRFRDIIVSAGFDEKCNDEFVKQPQKSH--HGEGVWKELFLRPTPPVRRML 895
Query: 318 FQKDIYSSIGWLPPAQDMNAIH----EVFKVARATT----LIALCGT----VPGYWFTVA 365
I + A + A+ +FK A TT L+A GT + +
Sbjct: 896 IAA---VGIHFFEHATGIEAVMLYGPRIFKKAGVTTKDRLLLATIGTGLTKITFLTISTF 1066
Query: 366 FIDVIGR---FAIQLMGFFF-MTVFMFALAIPYDHWTKRENRIGFLVMYAMTFF-FANFG 420
+D +GR I + G F +T+ F+L + K + ++ T+ F N G
Sbjct: 1067LLDRVGRRRLLQISVAGMIFGLTILGFSLTMVEYSSEKLVWALSLSIVATYTYVAFFNVG 1246
Query: 421 PNSTTFVVPAEIFPARLRSTCHGISSAA--GKAGAIIGAFGFLYASQSKDPKKRDAGYPA 478
T+V +EIFP RLR+ + I A G AI +F +Y +
Sbjct: 1247LAPVTWVYSSEIFPLRLRAQGNSIGVAVNRGMNAAISMSFISIYKA-------------- 1384
Query: 479 GIGMKNTLILLAVVNCLG-IFFTFLVPEANGKSLEEM 514
I + L A ++ + +FF F +PE GK+LEEM
Sbjct: 1385-ITIGGAFFLFAGMSVVAWVFFYFCLPETKGKALEEM 1492
>TC9159 similar to UP|HXT4_YEAST (P32467) Low-affinity glucose transporter
HXT4 (Low-affinity glucose transporter LGT1), partial
(5%)
Length = 728
Score = 51.6 bits (122), Expect = 3e-07
Identities = 33/116 (28%), Positives = 56/116 (47%), Gaps = 3/116 (2%)
Frame = +3
Query: 402 NRIGFLVMYAMTFFFANFGPNSTT--FVVPAEIFPARLRSTCHGISSAAGKAGAIIGAFG 459
+++GFL + + + F P T +VV +EI+P R R C GI+S ++ +
Sbjct: 102 SKLGFLALGGLALYIIFFSPGMGTVPWVVNSEIYPLRYRGICGGIASTTVWVSNLVVSQS 281
Query: 460 FLYASQSKDPKKRDAGYPAGIGMKNTLILLAVVNCLGIFFTFL-VPEANGKSLEEM 514
FL +Q+ IG T ++ ++ +GIFF + VPE G +EE+
Sbjct: 282 FLSLTQT-------------IGTAWTFMMFGIIAIIGIFFVLIFVPETKGVPMEEV 410
>BP039984
Length = 217
Score = 47.8 bits (112), Expect = 5e-06
Identities = 22/29 (75%), Positives = 24/29 (81%)
Frame = +3
Query: 1 MAREQLGVLTALDAAKTQWYHFTAIIIAG 29
MA+ + VL ALD AKTQWYHFTAIIIAG
Sbjct: 129 MAKGAIHVLNALDVAKTQWYHFTAIIIAG 215
>TC10135
Length = 1501
Score = 44.7 bits (104), Expect = 4e-05
Identities = 30/116 (25%), Positives = 53/116 (44%), Gaps = 3/116 (2%)
Frame = +2
Query: 402 NRIGFLVMYAMTFFFANFGPNSTT--FVVPAEIFPARLRSTCHGISSAAGKAGAIIGAFG 459
++ G+ + + + F P T +V+ +EI+P R R C GI+S +I +
Sbjct: 875 SKFGWAALVGLALYIITFSPGMGTVPWVINSEIYPFRYRGVCGGIASTTVWISNLIVSQS 1054
Query: 460 FLYASQSKDPKKRDAGYPAGIGMKNTLILLAVVNCLGIFFTFL-VPEANGKSLEEM 514
FL +Q+ IG T ++ ++ + IFF + VPE G +EE+
Sbjct: 1055FLSLTQT-------------IGTAWTFMMFGILAVVAIFFVIVFVPETKGVPMEEV 1183
>BP074036
Length = 405
Score = 40.4 bits (93), Expect = 7e-04
Identities = 30/101 (29%), Positives = 46/101 (44%), Gaps = 2/101 (1%)
Frame = -2
Query: 416 FANFGPNSTTFVVPAEIFPARLRSTCHGISSAAGKAG--AIIGAFGFLYASQSKDPKKRD 473
F + G T++ +EIFP LR+ + A + A++ +F +Y +
Sbjct: 404 FMSIGIGPVTWIYSSEIFPITLRAQGLAVCVAVNRIVNMAMLTSFISIYKA--------- 252
Query: 474 AGYPAGIGMKNTLILLAVVNCLGIFFTFLVPEANGKSLEEM 514
I M L LA VN L +F F +PE G+SLE+M
Sbjct: 251 ------ITMGGCLFALAGVNVLAFWFYFTLPETKGRSLEDM 147
>TC10748 similar to UP|Q7XA50 (Q7XA50) Sorbitol-like transporter, partial
(21%)
Length = 550
Score = 40.0 bits (92), Expect = 0.001
Identities = 32/111 (28%), Positives = 48/111 (42%), Gaps = 1/111 (0%)
Frame = +2
Query: 407 LVMYAMTFFFANFGPNSTTFVVPAEIFPARLRSTCHGISSAAGKAGAIIGAFGFLYASQS 466
++ Y TF + G T+V +EIFP RLR+ + + + G + S S
Sbjct: 5 VLSYVATF---SIGAGPITWVYSSEIFPLRLRAQGCAMGVVVNRVTS--GVISMTFLSLS 169
Query: 467 KDPKKRDAGYPAGIGMKNTLILLA-VVNCLGIFFTFLVPEANGKSLEEMSG 516
K GI + L + C IFF ++PE GK+LE+M G
Sbjct: 170 K-----------GITIGGAFFLFGGIAICGWIFFYTMLPETRGKTLEDMEG 289
>TC19595
Length = 500
Score = 37.7 bits (86), Expect = 0.005
Identities = 24/91 (26%), Positives = 41/91 (44%), Gaps = 1/91 (1%)
Frame = +1
Query: 426 FVVPAEIFPARLRSTCHGISSAAGKAGAIIGAFGFLYASQSKDPKKRDAGYPAGIGMKNT 485
+ V +EI+P R C G+S+ ++I + FL S + +G+ +
Sbjct: 10 WTVNSEIYPEEFRGVCGGMSATVNWVCSVIMSQSFLSISDA-------------VGLGGS 150
Query: 486 LILLAVVNCLGIFFTFL-VPEANGKSLEEMS 515
+L V+ + F L VPE G + EEM+
Sbjct: 151 FAILGVIAVVAFLFVLLFVPETKGLTFEEMT 243
>BG662351
Length = 407
Score = 37.7 bits (86), Expect = 0.005
Identities = 26/96 (27%), Positives = 42/96 (43%), Gaps = 1/96 (1%)
Frame = +1
Query: 420 GPNSTTFVVPAEIFPARLRSTCHGISSAAGKAGAIIGAFGFLYASQSKDPKKRDAGYPAG 479
G + +VV +EIFP ++ +++ GA + ++ F +
Sbjct: 130 GMGAVPWVVMSEIFPVNIKGQAGSLATLVNWFGAWLCSYTF--------------NFLMS 267
Query: 480 IGMKNTLILLAVVNCLGIF-FTFLVPEANGKSLEEM 514
T IL A +N LGI +VPE GKSLE++
Sbjct: 268 WSTYGTFILYAAINALGILSIVVVVPETKGKSLEQL 375
>TC12545 similar to UP|Q7XA50 (Q7XA50) Sorbitol-like transporter, partial
(18%)
Length = 482
Score = 35.4 bits (80), Expect = 0.024
Identities = 28/98 (28%), Positives = 42/98 (42%), Gaps = 1/98 (1%)
Frame = +2
Query: 420 GPNSTTFVVPAEIFPARLRSTCHGISSAAGKAGAIIGAFGFLYASQSKDPKKRDAGYPAG 479
G T+V +EIFP RLR+ I + + + + FL S +
Sbjct: 2 GSGPITWVYSSEIFPLRLRAQGVAIGAVVNRLTRGVISMTFLSLSNA------------- 142
Query: 480 IGMKNTLILLA-VVNCLGIFFTFLVPEANGKSLEEMSG 516
I + L A + C IF L+PE G++LEE+ G
Sbjct: 143 ITIGGAFFLFAGIAVCAWIFHYTLLPETRGQTLEEIEG 256
>TC12303 similar to GB|CAB10424.1|2245004|ATFCA6 membrane transporter like
protein {Arabidopsis thaliana;}, partial (25%)
Length = 545
Score = 33.9 bits (76), Expect = 0.069
Identities = 20/71 (28%), Positives = 35/71 (49%)
Frame = +3
Query: 72 AVNGVALCGTLAGQLFFGWLGDKMGRKKVYGLTLALMVFSSLASGLSFGHSAKGTIGTLC 131
A+ A+ G + G GW+ D+ GR+ +A+++ +L S +A L
Sbjct: 318 AIVSTAIAGAIIGAAVGGWINDRFGRR------VAIIIADTLFLIGSVIMAAAPNPAILL 479
Query: 132 FFRFWLGFGIG 142
F R ++GFG+G
Sbjct: 480 FGRVFVGFGVG 512
>TC9818 similar to UP|Q7XA50 (Q7XA50) Sorbitol-like transporter, partial
(62%)
Length = 1162
Score = 33.9 bits (76), Expect = 0.069
Identities = 41/171 (23%), Positives = 64/171 (36%)
Frame = +2
Query: 75 GVALCGTLAGQLFFGWLGDKMGRKKVYGLTLALMVFSSLASGLSFGHSAKGTIGTLCFFR 134
G+ +L G G D +GR+ A+ +L G S + L F R
Sbjct: 368 GIINLYSLIGSGLAGRTSDWIGRRYTIVFAGAIFFVGALLMGFSPNY------WFLMFGR 529
Query: 135 FWLGFGIGGDYPLSATIMSEYANKKTRGAFIASVFAMQGFGILAGGIVSLVVSTAFDHAF 194
F G GIG ++ +E + +RG + GIL G I + AF
Sbjct: 530 FIAGIGIGYALMIAPVYTAEVSPASSRGFLTSFPEVFINGGILLGYISNF--------AF 685
Query: 195 KAPPYKVDAAASLVPEADYVWRIILMFGAVPAGLTYYWRMKMPETARYTAL 245
KV WR++L GA+P+ + + MPE+ R+ +
Sbjct: 686 SKLSLKVG------------WRMMLGVGALPSVILGVGVLAMPESPRWLVM 802
>AV766904
Length = 611
Score = 32.3 bits (72), Expect = 0.20
Identities = 36/143 (25%), Positives = 57/143 (39%)
Frame = +3
Query: 84 GQLFFGWLGDKMGRKKVYGLTLALMVFSSLASGLSFGHSAKGTIGTLCFFRFWLGFGIGG 143
G G D +GR+ Y + LA ++F A + F + L F RF G GIG
Sbjct: 249 GSYIAGRTSDWIGRR--YTIVLAGLIFFVGAILMGFSPN----YAFLMFGRFVAGVGIGF 410
Query: 144 DYPLSATIMSEYANKKTRGAFIASVFAMQGFGILAGGIVSLVVSTAFDHAFKAPPYKVDA 203
+ ++ SE + +RG + GIL G I ++ F P ++
Sbjct: 411 AFLIAPVYTSEISPTLSRGFLTSLPEVFLNAGILIGYI--------SNYTFSKLPLRLG- 563
Query: 204 AASLVPEADYVWRIILMFGAVPA 226
WR++L GA+P+
Sbjct: 564 -----------WRLMLGIGAIPS 599
>TC12523
Length = 603
Score = 31.2 bits (69), Expect = 0.44
Identities = 11/37 (29%), Positives = 23/37 (61%)
Frame = -3
Query: 353 LCGTVPGYWFTVAFIDVIGRFAIQLMGFFFMTVFMFA 389
L GT +WFT+ + ++ F++ +GFF + +F+ +
Sbjct: 391 LIGTTITHWFTILLVFLLLIFSVTTLGFFKLLIFLLS 281
Database: LJGI
Posted date: Jul 30, 2004 11:16 AM
Number of letters in database: 14,692,800
Number of sequences in database: 28,460
Lambda K H
0.325 0.139 0.424
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 8,865,639
Number of Sequences: 28460
Number of extensions: 114485
Number of successful extensions: 834
Number of sequences better than 10.0: 50
Number of HSP's better than 10.0 without gapping: 816
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 825
length of query: 541
length of database: 4,897,600
effective HSP length: 95
effective length of query: 446
effective length of database: 2,193,900
effective search space: 978479400
effective search space used: 978479400
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 15 ( 7.0 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.6 bits)
S2: 57 (26.6 bits)
Medicago: description of AC148718.7


