

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
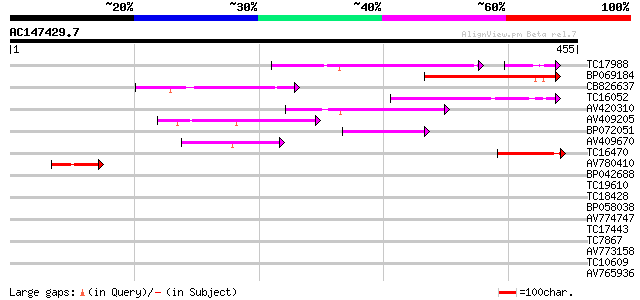
Query= AC147429.7 - phase: 0 /pseudo
(455 letters)
Database: LJGI
28,460 sequences; 14,692,800 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC17988 similar to UP|Q9FKP3 (Q9FKP3) Similarity to auxin-indepe... 109 2e-27
BP069184 105 2e-23
CB826637 86 1e-17
TC16052 homologue to UP|Q9LIN9 (Q9LIN9) Similarity to the auxin-... 79 2e-15
AV420310 74 7e-14
AV409205 66 1e-11
BP072051 52 2e-07
AV409670 49 1e-06
TC16470 weakly similar to UP|AAQ89634 (AAQ89634) At1g14020, part... 47 9e-06
AV780410 40 6e-04
BP042688 39 0.001
TC19610 31 0.37
TC18428 UP|APC2_CANFA (P12278) Apolipoprotein C-II precursor (Ap... 28 3.1
BP058038 28 4.1
AV774747 28 4.1
TC17443 27 5.3
TC7867 homologue to UP|CHSY_PUELO (P23569) Chalcone synthase (N... 27 7.0
AV773158 27 7.0
TC10609 27 7.0
AV765936 27 7.0
>TC17988 similar to UP|Q9FKP3 (Q9FKP3) Similarity to auxin-independent
growth promoter, partial (40%)
Length = 857
Score = 109 bits (273), Expect(2) = 2e-27
Identities = 65/175 (37%), Positives = 99/175 (56%), Gaps = 5/175 (2%)
Frame = +3
Query: 211 ENNTPYIALHLRYEKDMLAFTGCSHNLTKEETQELKKMRYSVKHWKEKEIDSK----SKR 266
+N ++ LHLR++KDM A + C K E L K R + W+ + ++S+ R
Sbjct: 63 KNAGKFVVLHLRFDKDMAAHSACDFGGGKAEKFALAKYRQVI--WQGRLLNSQFTDEELR 236
Query: 267 LKGSCPMTPREVAVFLEALGYPVDTKIYVAAGVIYGSEG-MKPLQKKFPNLLWHSSLATK 325
+G CPMTP EV + L ALG+ T++Y+A+ +YG E + L++ FP + SLA+
Sbjct: 237 SQGRCPMTPEEVGLLLAALGFDNSTRLYLASHKVYGGEARISTLRQLFPLMEDKKSLASS 416
Query: 326 EELQPFEGHLNQLAALDYYITVESDVFVYSYDGNMAKAARGHRKFDGFKKTISPD 380
E +G + LAALDYY+ + SD+F+ + GNM A GHR + KTI P+
Sbjct: 417 YERFQIKGKASLLAALDYYVGLHSDIFISASPGNMHNAVVGHRTYLNL-KTIRPN 578
Score = 29.3 bits (64), Expect(2) = 2e-27
Identities = 15/45 (33%), Positives = 23/45 (50%)
Frame = +1
Query: 398 WNDFSTKVKSIHAKKKGAPQARKIHRHPKFEETFYANPFPGCICQ 442
W++F V H ++G + RK PK ++ Y P P C+CQ
Sbjct: 622 WSEFEHAVVEGHQNRQGQLRLRK----PK--QSIYTYPAPDCMCQ 738
>BP069184
Length = 505
Score = 105 bits (261), Expect = 2e-23
Identities = 52/117 (44%), Positives = 74/117 (62%), Gaps = 8/117 (6%)
Frame = -3
Query: 334 HLNQLAALDYYITVESDVFVYSYDGNMAKAARGHRKFDGFKKTISPDKQRFVRLIDQLDN 393
H +Q+ ALDY +++ESDVF+ SY GNM +A GHR+F G +TISPD+Q RL D+LD
Sbjct: 503 HASQMPALDYIVSIESDVFIPSYSGNMDRAVEGHRRFLGRGRTISPDRQALARLFDKLDQ 324
Query: 394 G-LISWNDFSTKVKSIHAKKKGAPQARK-----IHRHPKF--EETFYANPFPGCICQ 442
G L + S ++ +H ++ G+P+ RK R +F EE FY NP P C+C+
Sbjct: 323 GTLTEGKELSNRIIDLHRRRLGSPRKRKGPISGTKRMDRFRSEEPFYVNPLPDCLCR 153
>CB826637
Length = 571
Score = 86.3 bits (212), Expect = 1e-17
Identities = 51/141 (36%), Positives = 75/141 (53%), Gaps = 10/141 (7%)
Frame = +3
Query: 102 FKEIFNWKNFVEVLNEDVQVVESLPP----------ELAAIKPALKAPVSWSKASYYRTD 151
F I++ ++F+ L DV +VES+P + I+P AP+SW Y TD
Sbjct: 6 FHGIYDVEHFIRTLRYDVNIVESIPESEKNGKKKKIKAFQIRPPRDAPISW-----YTTD 170
Query: 152 MLQLLKKHKVIKFTHTDSRLVNNGLASSIQRVRCRAMYEALRFAVPIEELGKKLVNRLRE 211
L+ +K+H I T RL Q +RCR Y ALRF I +L + +V++LR
Sbjct: 171 ALKKMKEHGAIYLTPFSHRLAEEIDIPEFQTLRCRVNYHALRFKPHIMKLSQSIVDKLRA 350
Query: 212 NNTPYIALHLRYEKDMLAFTG 232
P++++HLR+E DMLAF G
Sbjct: 351 QG-PFMSIHLRFEMDMLAFAG 410
>TC16052 homologue to UP|Q9LIN9 (Q9LIN9) Similarity to the auxin-independent
growth promoter, partial (24%)
Length = 809
Score = 79.0 bits (193), Expect = 2e-15
Identities = 47/138 (34%), Positives = 71/138 (51%), Gaps = 1/138 (0%)
Frame = +3
Query: 306 MKPLQKKFPNLLWHSSLATKEELQPFEGHLNQLAALDYYITVESDVFVYSYDGNMAKAAR 365
M PL+ FPNL+ LATK+EL F H+ LAALD+ + ++SDVFV + GN AK
Sbjct: 6 MAPLRNMFPNLVTKEELATKDELDGFRKHVTSLAALDFLVCLKSDVFVMTPGGNFAKLII 185
Query: 366 GHRKFDGFK-KTISPDKQRFVRLIDQLDNGLISWNDFSTKVKSIHAKKKGAPQARKIHRH 424
G R++ G + K+I PD+ + + + W F V H + G P+
Sbjct: 186 GARRYMGHRLKSIKPDQGLMSK---SFGDPYMGWATFVEDVVVTHQTRTGLPE----ETF 344
Query: 425 PKFEETFYANPFPGCICQ 442
P ++ + NP C+C+
Sbjct: 345 PNYD--LWENPLTPCMCK 392
>AV420310
Length = 408
Score = 73.6 bits (179), Expect = 7e-14
Identities = 47/139 (33%), Positives = 76/139 (53%), Gaps = 7/139 (5%)
Frame = +1
Query: 222 RYEKDMLAFTGCSHNLTKEETQELKKMRYSVKHWKEKEIDSKS------KRLKGSCPMTP 275
R EKD+ TGC L+ E + + R K K + ++S +++ G CP+T
Sbjct: 1 RMEKDVWVRTGCLPGLSPEFDEIVNNERIQ----KPKLLTARSNMTYHERKMAGLCPLTA 168
Query: 276 REVAVFLEALGYPVDTKIYVAAGV-IYGSEGMKPLQKKFPNLLWHSSLATKEELQPFEGH 334
EV L+ALG P +++IY A G + G E + PL ++FP+ LA EL+PF
Sbjct: 169 IEVTRLLKALGAPQNSRIYWAGGKPLGGKEALLPLIQEFPHFYNKEDLALPGELEPFAKK 348
Query: 335 LNQLAALDYYITVESDVFV 353
+ +AA+D+ ++ +SDVF+
Sbjct: 349 ASLMAAIDFIVSEKSDVFM 405
>AV409205
Length = 426
Score = 66.2 bits (160), Expect = 1e-11
Identities = 42/139 (30%), Positives = 71/139 (50%), Gaps = 8/139 (5%)
Frame = +3
Query: 119 VQVVESLPPELAAIK-----PALKAPVSWSKASYYRTDMLQLLKKHKVIKFTHTDSRLVN 173
V+V+ +LP +L + + P + P S S YY +L +LKKH V++ +D +
Sbjct: 3 VKVIRTLPKDLKSARRKKEIPVFRVPHSASPF-YYLHHVLPVLKKHSVVELVVSDGGCLQ 179
Query: 174 NGLASSI---QRVRCRAMYEALRFAVPIEELGKKLVNRLRENNTPYIALHLRYEKDMLAF 230
L S QR+RCR + AL+F ++EL +++ RLR P+IA ++ LA+
Sbjct: 180 AILPPSFKEYQRLRCRVSFHALQFRQEVQELSAQILQRLRAPGRPFIAFDPGMTRESLAY 359
Query: 231 TGCSHNLTKEETQELKKMR 249
GC+ T+ ++ R
Sbjct: 360 HGCAELFQDVHTELIQHKR 416
>BP072051
Length = 441
Score = 52.4 bits (124), Expect = 2e-07
Identities = 26/71 (36%), Positives = 40/71 (55%), Gaps = 1/71 (1%)
Frame = -3
Query: 268 KGSCPMTPREVAVFLEALGYPVDTKIYVAAGVIYGSEG-MKPLQKKFPNLLWHSSLATKE 326
+G CP+TP E + L ALG+ T IYVA +YG + L +P L+ +L + +
Sbjct: 436 EGLCPLTPEEAILMLAALGFNRKTHIYVAGSNLYGGRSRLVALTSLYPKLVTQENLLSPD 257
Query: 327 ELQPFEGHLNQ 337
EL+PF + +Q
Sbjct: 256 ELKPFANYSSQ 224
>AV409670
Length = 283
Score = 49.3 bits (116), Expect = 1e-06
Identities = 25/85 (29%), Positives = 46/85 (53%), Gaps = 3/85 (3%)
Frame = +2
Query: 139 PVSWSKASYYRTDMLQLLKKHKVIKFTHTDSRLVNNGLA---SSIQRVRCRAMYEALRFA 195
P S + ++Y ++L LKK KV+ + + L + IQR+RCR + +L+F
Sbjct: 20 PTSSASPNFYVKEVLPKLKKSKVVGIIIASGGALQSALPPTMAEIQRLRCRVAFHSLQFR 199
Query: 196 VPIEELGKKLVNRLRENNTPYIALH 220
I+ LG +++++LR P++ H
Sbjct: 200 PEIQMLGHRMIHKLRGLGQPFLVYH 274
>TC16470 weakly similar to UP|AAQ89634 (AAQ89634) At1g14020, partial (10%)
Length = 765
Score = 46.6 bits (109), Expect = 9e-06
Identities = 23/56 (41%), Positives = 36/56 (64%), Gaps = 1/56 (1%)
Frame = +3
Query: 392 DNGLISWNDFSTKVKSIHAKKKGAPQAR-KIHRHPKFEETFYANPFPGCICQKPTH 446
+NG+++W++FS+ VK HA + G+P R I PK E+ FYANP C +P++
Sbjct: 12 ENGVLNWDEFSSAVKESHADRMGSPSKRLAILDKPKEEDYFYANPHE---CLEPSN 170
>AV780410
Length = 548
Score = 40.4 bits (93), Expect = 6e-04
Identities = 21/42 (50%), Positives = 26/42 (61%)
Frame = +2
Query: 34 IHLCITMKNFYYIGQGNNTNGYLLVHANGGLNQMKTGISDMV 75
+H C+ Y QG + YL V +NGGLN M+TGISDMV
Sbjct: 428 LHQCVKPTARYNGAQG--VDRYLTVRSNGGLNHMRTGISDMV 547
>BP042688
Length = 480
Score = 39.3 bits (90), Expect = 0.001
Identities = 21/53 (39%), Positives = 31/53 (57%), Gaps = 2/53 (3%)
Frame = -1
Query: 349 SDVFVYSYDGNMAKAARGHRKF--DGFKKTISPDKQRFVRLIDQLDNGLISWN 399
S+VFV + GN GHR+F DG KTI PDK++ V L + + L +++
Sbjct: 480 SEVFVTTQGGNFPHFLMGHRRFLYDGHAKTIIPDKRKLVVLFEDMSIRLCAFS 322
>TC19610
Length = 513
Score = 31.2 bits (69), Expect = 0.37
Identities = 13/32 (40%), Positives = 17/32 (52%)
Frame = +3
Query: 411 KKKGAPQARKIHRHPKFEETFYANPFPGCICQ 442
K+KG K + EE FY NP P C+C+
Sbjct: 273 KRKGPISGTKRMDRFRSEEPFYVNPLPDCLCR 368
>TC18428 UP|APC2_CANFA (P12278) Apolipoprotein C-II precursor (Apo-CII),
partial (8%)
Length = 512
Score = 28.1 bits (61), Expect = 3.1
Identities = 11/32 (34%), Positives = 20/32 (62%)
Frame = -2
Query: 23 VKFDISDSHSIIHLCITMKNFYYIGQGNNTNG 54
+K +++H+IIH T+ NF++ G N +G
Sbjct: 490 IKL*ENNTHNIIHPQTTLPNFFFFNSGINRSG 395
>BP058038
Length = 552
Score = 27.7 bits (60), Expect = 4.1
Identities = 16/39 (41%), Positives = 18/39 (46%)
Frame = +1
Query: 220 HLRYEKDMLAFTGCSHNLTKEETQELKKMRYSVKHWKEK 258
H Y D +T C NL EE Q L R + K KEK
Sbjct: 13 HFYYSADTYIYTNC*CNLVYEEKQVLLLRRCTGKCCKEK 129
>AV774747
Length = 468
Score = 27.7 bits (60), Expect = 4.1
Identities = 12/35 (34%), Positives = 22/35 (62%), Gaps = 3/35 (8%)
Frame = +1
Query: 185 CRAMYEALRFAVPIEELGK---KLVNRLRENNTPY 216
C +Y ++ + +E L + +L+NRL EN+TP+
Sbjct: 331 CELVYRSIPVSPQVESLSRYVLQLMNRLGENHTPF 435
>TC17443
Length = 531
Score = 27.3 bits (59), Expect = 5.3
Identities = 14/39 (35%), Positives = 21/39 (52%)
Frame = +2
Query: 341 LDYYITVESDVFVYSYDGNMAKAARGHRKFDGFKKTISP 379
+D+YI+ ESDVFV + G + G R G + + P
Sbjct: 110 IDFYISSESDVFVPAISGLLYANVAGKRIGSGKTQILVP 226
>TC7867 homologue to UP|CHSY_PUELO (P23569) Chalcone synthase
(Naringenin-chalcone synthase) , complete
Length = 1387
Score = 26.9 bits (58), Expect = 7.0
Identities = 12/43 (27%), Positives = 22/43 (50%)
Frame = +3
Query: 296 AAGVIYGSEGMKPLQKKFPNLLWHSSLATKEELQPFEGHLNQL 338
AA VI GS+ + ++K L+W + + +GHL ++
Sbjct: 708 AAAVIVGSDPVPEIEKPLFELVWTAQTIAPDSDGAIDGHLREV 836
>AV773158
Length = 490
Score = 26.9 bits (58), Expect = 7.0
Identities = 12/32 (37%), Positives = 18/32 (55%)
Frame = -1
Query: 237 LTKEETQELKKMRYSVKHWKEKEIDSKSKRLK 268
L K+ETQ +K + K W + SK+K L+
Sbjct: 256 LRKQETQRVKVVNRRTKTWDNESKGSKTKNLR 161
>TC10609
Length = 529
Score = 26.9 bits (58), Expect = 7.0
Identities = 10/22 (45%), Positives = 14/22 (63%)
Frame = -2
Query: 215 PYIALHLRYEKDMLAFTGCSHN 236
PY+ + LR+ D+ F G SHN
Sbjct: 225 PYLMIMLRFSNDLQHFYGISHN 160
>AV765936
Length = 447
Score = 26.9 bits (58), Expect = 7.0
Identities = 13/43 (30%), Positives = 25/43 (57%), Gaps = 2/43 (4%)
Frame = +3
Query: 85 LVFPTLDHNSFWTDPSD--FKEIFNWKNFVEVLNEDVQVVESL 125
L+ T + +F+ D D +I NW+NF V++ V++ +S+
Sbjct: 150 LILKTR*YQTFYIDHWDNPIIDILNWENFESVVSRIVKIYDSI 278
Database: LJGI
Posted date: Jul 30, 2004 11:16 AM
Number of letters in database: 14,692,800
Number of sequences in database: 28,460
Lambda K H
0.320 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 7,912,215
Number of Sequences: 28460
Number of extensions: 104729
Number of successful extensions: 540
Number of sequences better than 10.0: 43
Number of HSP's better than 10.0 without gapping: 531
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 532
length of query: 455
length of database: 4,897,600
effective HSP length: 93
effective length of query: 362
effective length of database: 2,250,820
effective search space: 814796840
effective search space used: 814796840
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 57 (26.6 bits)
Medicago: description of AC147429.7


