

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146865.2 - phase: 0
(107 letters)
Database: LJGI
28,460 sequences; 14,692,800 total letters
Searching..................................................done
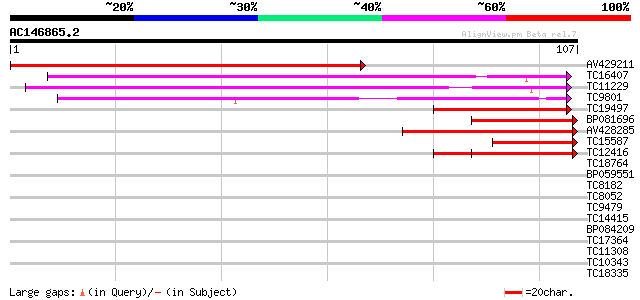
Score E
Sequences producing significant alignments: (bits) Value
AV429211 136 6e-34
TC16407 similar to GB|AAL06810.1|15809764|AY054149 At1g76300/F15... 57 4e-10
TC11229 homologue to UP|Q9LM92 (Q9LM92) F2D10.7 (At1g20580/F2D10... 56 8e-10
TC9801 homologue to UP|Q8LAC6 (Q8LAC6) Glycine rich protein-like... 54 3e-09
TC19497 weakly similar to UP|Q8LA02 (Q8LA02) Contains similarity... 42 2e-05
BP081696 40 5e-05
AV428285 40 6e-05
TC15587 similar to UP|Q9LHG0 (Q9LHG0) Genomic DNA, chromosome 3,... 39 2e-04
TC12416 similar to GB|AAH56066.1|33417132|BC056066 sop-prov prot... 38 3e-04
TC18764 similar to UP|Q8LBJ5 (Q8LBJ5) Nucleic acid binding prote... 35 0.002
BP059551 34 0.003
TC8182 weakly similar to UP|Q93VA8 (Q93VA8) At1g76010/T4O12_22, ... 33 0.007
TC8052 similar to GB|AAP13431.1|30023796|BT006323 At5g02530 {Ara... 33 0.007
TC9479 similar to AAQ56832 (AAQ56832) At2g34040, partial (12%) 31 0.028
TC14415 similar to UP|O04237 (O04237) Transcription factor, part... 30 0.047
BP084209 30 0.062
TC17364 homologue to UP|Q9VVG2 (Q9VVG2) CG13731-PA, partial (3%) 30 0.080
TC11308 similar to UP|CENB_HUMAN (P07199) Major centromere autoa... 30 0.080
TC10343 similar to UP|Q9SZZ1 (Q9SZZ1) Fibrillarin-like protein (... 28 0.18
TC18335 similar to UP|Q7XJ13 (Q7XJ13) Chloroplast nucleoid DNA-b... 24 3.4
>AV429211
Length = 304
Score = 136 bits (342), Expect = 6e-34
Identities = 67/67 (100%), Positives = 67/67 (100%)
Frame = +2
Query: 1 MKLNNETVSIELKNGTIVHGTITGVDISMNTHLKTVKLTLKGKNPVTLDHLSVRGNNIRY 60
MKLNNETVSIELKNGTIVHGTITGVDISMNTHLKTVKLTLKGKNPVTLDHLSVRGNNIRY
Sbjct: 104 MKLNNETVSIELKNGTIVHGTITGVDISMNTHLKTVKLTLKGKNPVTLDHLSVRGNNIRY 283
Query: 61 YILPDSL 67
YILPDSL
Sbjct: 284 YILPDSL 304
>TC16407 similar to GB|AAL06810.1|15809764|AY054149 At1g76300/F15M4_20
{Arabidopsis thaliana;}, partial (92%)
Length = 678
Score = 57.4 bits (137), Expect = 4e-10
Identities = 32/102 (31%), Positives = 55/102 (53%), Gaps = 3/102 (2%)
Frame = +3
Query: 8 VSIELKNGTIVHGTITGVDISMNTHLKTVKLTLKGKNPVTLDHLSVRGNNIRYYILPDSL 67
VS+ELK+G + G++ + + N L+++ T K L+H+ +RG+ +R+ ++PD L
Sbjct: 108 VSVELKSGELYRGSMIECEDNWNCQLESITYTAKDGKTSQLEHVFIRGSKVRFMVIPDML 287
Query: 68 NLETLLVEEAPRVKPKKPTAGKPLGRGRG---RGRGRGRGRG 106
+ RVK K + G +GRGR R + + GRG
Sbjct: 288 KNAPMFKRLDARVKGKGASLG--VGRGRAVAMRAKAQAAGRG 407
>TC11229 homologue to UP|Q9LM92 (Q9LM92) F2D10.7 (At1g20580/F2D10_6),
partial (80%)
Length = 591
Score = 56.2 bits (134), Expect = 8e-10
Identities = 33/114 (28%), Positives = 55/114 (47%), Gaps = 11/114 (9%)
Frame = +2
Query: 4 NNETVSIELKNGTIVHGTITGVDISMNTHLKTVKLTLKGKNPVTLDHLSVRGNNIRYYIL 63
+ V++ELK+G + G + + + N L+++ T K L+H+ +RG+ +R+ ++
Sbjct: 68 SGHVVTVELKSGELYRGNMIECEDNWNCQLESITYTAKDGKTSQLEHVFIRGSKVRFMVI 247
Query: 64 PDSLNLETLLVEEAPRVKPKKPTAGKPLGRGRGR-----------GRGRGRGRG 106
PD L + ++K K G LG GRGR GRG GRG
Sbjct: 248 PDMLKNAPMFKRLDAKIKGK----GASLGVGRGRAVAMRAKAQSAGRGAAPGRG 397
>TC9801 homologue to UP|Q8LAC6 (Q8LAC6) Glycine rich protein-like, complete
Length = 981
Score = 54.3 bits (129), Expect = 3e-09
Identities = 38/98 (38%), Positives = 49/98 (49%), Gaps = 1/98 (1%)
Frame = +2
Query: 10 IELKNGTIVHGTITGVDISMNTHLKTVKLTLK-GKNPVTLDHLSVRGNNIRYYILPDSLN 68
+ELKNG +G + D MN HL+ V T K G + +RGN I+Y +PD
Sbjct: 146 VELKNGETYNGHLVNCDTWMNIHLREVICTNKDGDRFWRMPECYIRGNTIKYLRVPDE-- 319
Query: 69 LETLLVEEAPRVKPKKPTAGKPLGRGRGRGRGRGRGRG 106
V + + + K T KP G GRGRGRG GR G
Sbjct: 320 -----VIDKVQEETKSRTDRKPPGVGRGRGRG-GREEG 415
>TC19497 weakly similar to UP|Q8LA02 (Q8LA02) Contains similarity to
RNA-binding protein from Arabidopsis thaliana gi|2129727
and contains RNA recognition PF|00076 domain, partial
(11%)
Length = 436
Score = 41.6 bits (96), Expect = 2e-05
Identities = 18/26 (69%), Positives = 20/26 (76%)
Frame = +2
Query: 81 KPKKPTAGKPLGRGRGRGRGRGRGRG 106
+ + P G LGRGRGRGRGRGRGRG
Sbjct: 95 RERTPREGGGLGRGRGRGRGRGRGRG 172
Score = 35.8 bits (81), Expect = 0.001
Identities = 15/16 (93%), Positives = 15/16 (93%)
Frame = +2
Query: 92 GRGRGRGRGRGRGRGR 107
G GRGRGRGRGRGRGR
Sbjct: 122 GLGRGRGRGRGRGRGR 169
Score = 34.7 bits (78), Expect = 0.003
Identities = 18/30 (60%), Positives = 20/30 (66%), Gaps = 1/30 (3%)
Frame = +2
Query: 79 RVKPKKPTAGKPLGRGRGRGRGRGRG-RGR 107
R + + G GRGRGRGRGRGRG RGR
Sbjct: 95 RERTPREGGGLGRGRGRGRGRGRGRGFRGR 184
>BP081696
Length = 383
Score = 40.4 bits (93), Expect = 5e-05
Identities = 17/20 (85%), Positives = 18/20 (90%)
Frame = -3
Query: 88 GKPLGRGRGRGRGRGRGRGR 107
G+ GRGRGRGRGRGRGRGR
Sbjct: 192 GRSRGRGRGRGRGRGRGRGR 133
>AV428285
Length = 239
Score = 40.0 bits (92), Expect = 6e-05
Identities = 20/33 (60%), Positives = 22/33 (66%)
Frame = +3
Query: 75 EEAPRVKPKKPTAGKPLGRGRGRGRGRGRGRGR 107
EEA R K + G+GRGRGRGRGRGRGR
Sbjct: 51 EEAKRSKMLELGHTSSTGKGRGRGRGRGRGRGR 149
>TC15587 similar to UP|Q9LHG0 (Q9LHG0) Genomic DNA, chromosome 3, P1 clone:
MQC3 (At3g12480/MQC3.32), partial (7%)
Length = 768
Score = 38.5 bits (88), Expect = 2e-04
Identities = 16/16 (100%), Positives = 16/16 (100%)
Frame = +2
Query: 92 GRGRGRGRGRGRGRGR 107
GRGRGRGRGRGRGRGR
Sbjct: 29 GRGRGRGRGRGRGRGR 76
Score = 32.0 bits (71), Expect = 0.016
Identities = 13/18 (72%), Positives = 15/18 (83%)
Frame = +2
Query: 86 TAGKPLGRGRGRGRGRGR 103
+ G+ GRGRGRGRGRGR
Sbjct: 23 STGRGRGRGRGRGRGRGR 76
>TC12416 similar to GB|AAH56066.1|33417132|BC056066 sop-prov protein
{Xenopus laevis;} , partial (32%)
Length = 416
Score = 37.7 bits (86), Expect = 3e-04
Identities = 16/20 (80%), Positives = 17/20 (85%)
Frame = +3
Query: 88 GKPLGRGRGRGRGRGRGRGR 107
G+ GRG GRGRGRGRGRGR
Sbjct: 153 GEGRGRGEGRGRGRGRGRGR 212
Score = 37.7 bits (86), Expect = 3e-04
Identities = 16/20 (80%), Positives = 17/20 (85%)
Frame = +3
Query: 88 GKPLGRGRGRGRGRGRGRGR 107
G+ GRGRG GRGRGRGRGR
Sbjct: 147 GRGEGRGRGEGRGRGRGRGR 206
Score = 36.6 bits (83), Expect = 7e-04
Identities = 16/27 (59%), Positives = 17/27 (62%)
Frame = +3
Query: 81 KPKKPTAGKPLGRGRGRGRGRGRGRGR 107
+P G GRGRG GRGRG GRGR
Sbjct: 108 EPSAEGFGDGFGRGRGEGRGRGEGRGR 188
Score = 36.2 bits (82), Expect = 9e-04
Identities = 15/20 (75%), Positives = 16/20 (80%)
Frame = +3
Query: 88 GKPLGRGRGRGRGRGRGRGR 107
G+ G GRGRG GRGRGRGR
Sbjct: 141 GRGRGEGRGRGEGRGRGRGR 200
Score = 28.9 bits (63), Expect = 0.14
Identities = 13/19 (68%), Positives = 14/19 (73%)
Frame = +3
Query: 88 GKPLGRGRGRGRGRGRGRG 106
G+ GRGRGRGRGR RG
Sbjct: 171 GEGRGRGRGRGRGRRGRRG 227
Score = 28.5 bits (62), Expect = 0.18
Identities = 12/21 (57%), Positives = 14/21 (66%)
Frame = +3
Query: 87 AGKPLGRGRGRGRGRGRGRGR 107
+G+P G G G GRGRG GR
Sbjct: 102 SGEPSAEGFGDGFGRGRGEGR 164
Score = 26.6 bits (57), Expect = 0.68
Identities = 12/19 (63%), Positives = 13/19 (68%)
Frame = +3
Query: 88 GKPLGRGRGRGRGRGRGRG 106
G+ GRGRGRGR RG G
Sbjct: 177 GRGRGRGRGRGRRGRRGDG 233
>TC18764 similar to UP|Q8LBJ5 (Q8LBJ5) Nucleic acid binding protein-like,
partial (35%)
Length = 848
Score = 35.0 bits (79), Expect = 0.002
Identities = 15/16 (93%), Positives = 15/16 (93%)
Frame = +3
Query: 92 GRGRGRGRGRGRGRGR 107
GRGRGRGRGRGRGR R
Sbjct: 174 GRGRGRGRGRGRGRFR 221
Score = 29.6 bits (65), Expect = 0.080
Identities = 15/20 (75%), Positives = 16/20 (80%)
Frame = +3
Query: 88 GKPLGRGRGRGRGRGRGRGR 107
G+ GRGRGRGRGR R RGR
Sbjct: 174 GRGRGRGRGRGRGRFR-RGR 230
>BP059551
Length = 454
Score = 34.3 bits (77), Expect = 0.003
Identities = 15/18 (83%), Positives = 15/18 (83%)
Frame = -1
Query: 90 PLGRGRGRGRGRGRGRGR 107
P GRGRGRGRG RGRGR
Sbjct: 358 PQGRGRGRGRGGYRGRGR 305
Score = 30.4 bits (67), Expect = 0.047
Identities = 13/15 (86%), Positives = 13/15 (86%)
Frame = -1
Query: 92 GRGRGRGRGRGRGRG 106
GRGRGRG RGRGRG
Sbjct: 346 GRGRGRGGYRGRGRG 302
Score = 27.7 bits (60), Expect = 0.31
Identities = 12/17 (70%), Positives = 13/17 (75%)
Frame = -1
Query: 88 GKPLGRGRGRGRGRGRG 104
G+ GRGRG RGRGRG
Sbjct: 352 GRGRGRGRGGYRGRGRG 302
>TC8182 weakly similar to UP|Q93VA8 (Q93VA8) At1g76010/T4O12_22, partial
(13%)
Length = 611
Score = 33.1 bits (74), Expect = 0.007
Identities = 16/19 (84%), Positives = 16/19 (84%), Gaps = 1/19 (5%)
Frame = +3
Query: 90 PLGRGRGRGR-GRGRGRGR 107
P RGRGRGR GRGRGRGR
Sbjct: 195 PAPRGRGRGRGGRGRGRGR 251
Score = 30.8 bits (68), Expect = 0.036
Identities = 15/17 (88%), Positives = 15/17 (88%), Gaps = 1/17 (5%)
Frame = +3
Query: 92 GRGRGRGRGRGRG-RGR 107
GRGRG GRGRGRG RGR
Sbjct: 81 GRGRGFGRGRGRGFRGR 131
Score = 30.4 bits (67), Expect = 0.047
Identities = 15/20 (75%), Positives = 16/20 (80%), Gaps = 1/20 (5%)
Frame = +3
Query: 88 GKPLGRGRGRGRG-RGRGRG 106
G+ G GRGRGRG RGRGRG
Sbjct: 81 GRGRGFGRGRGRGFRGRGRG 140
Score = 23.5 bits (49), Expect = 5.8
Identities = 12/20 (60%), Positives = 13/20 (65%), Gaps = 1/20 (5%)
Frame = +3
Query: 88 GKPLGRG-RGRGRGRGRGRG 106
G+ GRG RGRGRGR G
Sbjct: 207 GRGRGRGGRGRGRGRNASWG 266
>TC8052 similar to GB|AAP13431.1|30023796|BT006323 At5g02530 {Arabidopsis
thaliana;}, partial (55%)
Length = 1389
Score = 33.1 bits (74), Expect = 0.007
Identities = 14/20 (70%), Positives = 15/20 (75%)
Frame = +2
Query: 88 GKPLGRGRGRGRGRGRGRGR 107
G+ G RGRGRGRG GRGR
Sbjct: 758 GRGRGHRRGRGRGRGHGRGR 817
Score = 32.7 bits (73), Expect = 0.010
Identities = 14/19 (73%), Positives = 15/19 (78%)
Frame = +2
Query: 88 GKPLGRGRGRGRGRGRGRG 106
G+ RGRGRGRG GRGRG
Sbjct: 764 GRGHRRGRGRGRGHGRGRG 820
Score = 25.4 bits (54), Expect = 1.5
Identities = 11/16 (68%), Positives = 11/16 (68%)
Frame = +2
Query: 92 GRGRGRGRGRGRGRGR 107
G GRGRG RGRGR
Sbjct: 746 GGDGGRGRGHRRGRGR 793
>TC9479 similar to AAQ56832 (AAQ56832) At2g34040, partial (12%)
Length = 707
Score = 31.2 bits (69), Expect = 0.028
Identities = 15/22 (68%), Positives = 16/22 (72%), Gaps = 3/22 (13%)
Frame = +1
Query: 88 GKPLGRGRGRG---RGRGRGRG 106
G+ RGRGRG RGRGRGRG
Sbjct: 337 GRGFARGRGRGWGGRGRGRGRG 402
Score = 25.4 bits (54), Expect = 1.5
Identities = 11/15 (73%), Positives = 11/15 (73%)
Frame = +1
Query: 92 GRGRGRGRGRGRGRG 106
G GRG RGRGRG G
Sbjct: 331 GGGRGFARGRGRGWG 375
>TC14415 similar to UP|O04237 (O04237) Transcription factor, partial (83%)
Length = 1666
Score = 30.4 bits (67), Expect = 0.047
Identities = 16/28 (57%), Positives = 18/28 (64%), Gaps = 1/28 (3%)
Frame = +2
Query: 80 VKPKKPTAGKPLGRGRGRGRG-RGRGRG 106
+KP + P GRGRGRGRG RG G G
Sbjct: 1166 LKPAEGETYYPSGRGRGRGRGSRGGGGG 1249
Score = 29.6 bits (65), Expect = 0.080
Identities = 18/44 (40%), Positives = 23/44 (51%)
Frame = +2
Query: 64 PDSLNLETLLVEEAPRVKPKKPTAGKPLGRGRGRGRGRGRGRGR 107
P + + L +A R +P+ G GRG RG GRG GRGR
Sbjct: 374 PAQMPSKPLPPAQAVRDARNEPSRG---GRGGARGAGRGFGRGR 496
Score = 26.6 bits (57), Expect = 0.68
Identities = 13/27 (48%), Positives = 15/27 (55%)
Frame = +2
Query: 79 RVKPKKPTAGKPLGRGRGRGRGRGRGR 105
R +P + G G GRG GRGRG R
Sbjct: 428 RNEPSRGGRGGARGAGRGFGRGRGFNR 508
>BP084209
Length = 502
Score = 30.0 bits (66), Expect = 0.062
Identities = 16/24 (66%), Positives = 16/24 (66%), Gaps = 1/24 (4%)
Frame = -3
Query: 84 KPTAGKPLGRGRGRG-RGRGRGRG 106
KP GRGRGRG RGRGRG G
Sbjct: 389 KPHRSNFNGRGRGRGGRGRGRGGG 318
>TC17364 homologue to UP|Q9VVG2 (Q9VVG2) CG13731-PA, partial (3%)
Length = 446
Score = 29.6 bits (65), Expect = 0.080
Identities = 14/22 (63%), Positives = 15/22 (67%)
Frame = -1
Query: 86 TAGKPLGRGRGRGRGRGRGRGR 107
T G+ GR RGR RGR RGR R
Sbjct: 443 TRGRTRGRTRGRTRGRTRGRTR 378
Score = 26.6 bits (57), Expect = 0.68
Identities = 12/19 (63%), Positives = 13/19 (68%)
Frame = -1
Query: 86 TAGKPLGRGRGRGRGRGRG 104
T G+ GR RGR RGR RG
Sbjct: 431 TRGRTRGRTRGRTRGRTRG 375
Score = 23.9 bits (50), Expect = 4.4
Identities = 11/15 (73%), Positives = 11/15 (73%)
Frame = -1
Query: 93 RGRGRGRGRGRGRGR 107
R RGR RGR RGR R
Sbjct: 446 RTRGRTRGRTRGRTR 402
>TC11308 similar to UP|CENB_HUMAN (P07199) Major centromere autoantigen B
(Centromere protein B) (CENP-B), partial (4%)
Length = 756
Score = 29.6 bits (65), Expect = 0.080
Identities = 13/16 (81%), Positives = 13/16 (81%)
Frame = -1
Query: 92 GRGRGRGRGRGRGRGR 107
GRG G GRGRGR RGR
Sbjct: 300 GRGGGSGRGRGR*RGR 253
Score = 27.7 bits (60), Expect = 0.31
Identities = 12/15 (80%), Positives = 12/15 (80%)
Frame = -1
Query: 92 GRGRGRGRGRGRGRG 106
G G GRGRGR RGRG
Sbjct: 294 GGGSGRGRGR*RGRG 250
Score = 26.9 bits (58), Expect = 0.52
Identities = 12/17 (70%), Positives = 13/17 (75%)
Frame = -1
Query: 88 GKPLGRGRGRGRGRGRG 104
G+ G GRGRGR RGRG
Sbjct: 300 GRGGGSGRGRGR*RGRG 250
>TC10343 similar to UP|Q9SZZ1 (Q9SZZ1) Fibrillarin-like protein (Fibrillarin
2) (AtFib2), partial (85%)
Length = 1116
Score = 28.5 bits (62), Expect = 0.18
Identities = 12/15 (80%), Positives = 12/15 (80%)
Frame = +1
Query: 92 GRGRGRGRGRGRGRG 106
GRGRG G GRG GRG
Sbjct: 91 GRGRGDGGGRGGGRG 135
Score = 28.1 bits (61), Expect = 0.23
Identities = 12/15 (80%), Positives = 12/15 (80%)
Frame = +1
Query: 93 RGRGRGRGRGRGRGR 107
RGRGRG G GRG GR
Sbjct: 88 RGRGRGDGGGRGGGR 132
Score = 24.3 bits (51), Expect = 3.4
Identities = 14/23 (60%), Positives = 14/23 (60%), Gaps = 1/23 (4%)
Frame = +1
Query: 86 TAGKPLGRGRGRGRGR-GRGRGR 107
T K G GRG G GR GRG GR
Sbjct: 136 TPFKARGGGRGGGGGRGGRGGGR 204
Score = 23.1 bits (48), Expect = 7.5
Identities = 13/24 (54%), Positives = 14/24 (58%), Gaps = 5/24 (20%)
Frame = +1
Query: 88 GKPLGRG-----RGRGRGRGRGRG 106
G+ GRG RG GRG G GRG
Sbjct: 115 GRGGGRGTPFKARGGGRGGGGGRG 186
Score = 23.1 bits (48), Expect = 7.5
Identities = 13/25 (52%), Positives = 14/25 (56%), Gaps = 5/25 (20%)
Frame = +1
Query: 88 GKPLGRGRGRGRGRG-----RGRGR 107
G+ G G GRG GRG RG GR
Sbjct: 91 GRGRGDGGGRGGGRGTPFKARGGGR 165
>TC18335 similar to UP|Q7XJ13 (Q7XJ13) Chloroplast nucleoid DNA-binding
protein-like protein, partial (19%)
Length = 588
Score = 24.3 bits (51), Expect = 3.4
Identities = 14/32 (43%), Positives = 17/32 (52%), Gaps = 4/32 (12%)
Frame = +3
Query: 77 APRVKP----KKPTAGKPLGRGRGRGRGRGRG 104
A +VKP ++ P GRGRGRGRG
Sbjct: 393 ADQVKPLNEYEEDGEXSPXMPXXGRGRGRGRG 488
Score = 23.9 bits (50), Expect = 4.4
Identities = 9/9 (100%), Positives = 9/9 (100%)
Frame = +3
Query: 98 GRGRGRGRG 106
GRGRGRGRG
Sbjct: 462 GRGRGRGRG 488
Score = 23.5 bits (49), Expect = 5.8
Identities = 10/22 (45%), Positives = 11/22 (49%)
Frame = +3
Query: 75 EEAPRVKPKKPTAGKPLGRGRG 96
EE P P G+ GRGRG
Sbjct: 423 EEDGEXSPXMPXXGRGRGRGRG 488
Database: LJGI
Posted date: Jul 30, 2004 11:16 AM
Number of letters in database: 14,692,800
Number of sequences in database: 28,460
Lambda K H
0.315 0.138 0.394
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 1,543,017
Number of Sequences: 28460
Number of extensions: 19558
Number of successful extensions: 611
Number of sequences better than 10.0: 143
Number of HSP's better than 10.0 without gapping: 227
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 419
length of query: 107
length of database: 4,897,600
effective HSP length: 83
effective length of query: 24
effective length of database: 2,535,420
effective search space: 60850080
effective search space used: 60850080
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.9 bits)
S2: 47 (22.7 bits)
Medicago: description of AC146865.2


