

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146752.2 + phase: 0
(394 letters)
Database: LJGI
28,460 sequences; 14,692,800 total letters
Searching..................................................done
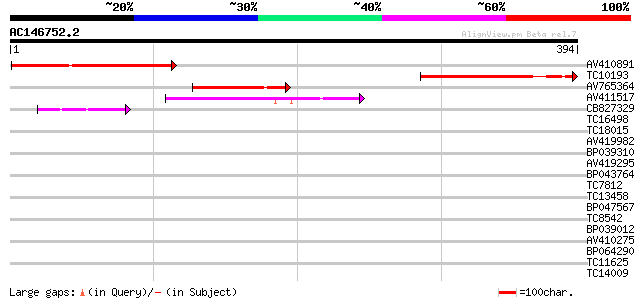
Score E
Sequences producing significant alignments: (bits) Value
AV410891 170 3e-43
TC10193 similar to UP|Q944L7 (Q944L7) At1g18480/F15H18_1, partia... 139 8e-34
AV765364 99 9e-22
AV411517 75 2e-14
CB827329 44 6e-05
TC16498 similar to GB|AAP37835.1|30725626|BT008476 At1g07010 {Ar... 37 0.006
TC18015 33 0.082
AV419982 33 0.11
BP039310 33 0.11
AV419295 33 0.11
BP043764 32 0.14
TC7812 UP|Q9LKJ6 (Q9LKJ6) Water-selective transport intrinsic me... 32 0.18
TC13458 weakly similar to UP|Z282_HUMAN (Q9UDV7) Zinc finger pro... 32 0.24
BP047567 31 0.31
TC8542 similar to UP|RK1_PEA (P49208) 50S ribosomal protein L1, ... 31 0.31
BP039012 26 0.32
AV410275 31 0.41
BP064290 31 0.41
TC11625 weakly similar to UP|O81015 (O81015) At2g26920 protein, ... 31 0.41
TC14009 similar to UP|HSF5_ARATH (Q9S7U5) Heat shock factor prot... 31 0.41
>AV410891
Length = 380
Score = 170 bits (431), Expect = 3e-43
Identities = 84/115 (73%), Positives = 94/115 (81%)
Frame = +3
Query: 2 EFEKQNSNTFCNQIPNFLSSFIDTFVDFSVSGGLFLPPPPSSPPPIPTRLPSPSRLIAIG 61
E E S +FC+ +PN LSSF+DTFVDFSVSG LPPPP P P+PTRLPS SRLIAIG
Sbjct: 42 EEETSTSTSFCHHVPNLLSSFVDTFVDFSVSGLFLLPPPP--PSPLPTRLPSQSRLIAIG 215
Query: 62 DLHGDLKKSKEALSIAGLIDSSGNYTGGSATVVQIGDVLDRGGDEIKILYLLEKL 116
DLHGDL +SK+AL +AGLIDSS Y GGSATVVQ+GDV DRGGDE+KILY LEKL
Sbjct: 216 DLHGDLDRSKQALRLAGLIDSSDRYAGGSATVVQVGDVFDRGGDELKILYFLEKL 380
>TC10193 similar to UP|Q944L7 (Q944L7) At1g18480/F15H18_1, partial (22%)
Length = 793
Score = 139 bits (350), Expect = 8e-34
Identities = 74/110 (67%), Positives = 86/110 (77%), Gaps = 1/110 (0%)
Frame = +3
Query: 286 NCDCSSLEHVLSTIPGVKRMIMGHTIQKEGINGVCENKAIRIDVGMSKGCGGGLPEVLEI 345
+CDCS+LE+VLST+PGVKRM+MGHTIQ GINGVC+N+AIRIDVGMSKGCGGGLPEVLEI
Sbjct: 15 DCDCSTLEYVLSTVPGVKRMVMGHTIQTVGINGVCDNRAIRIDVGMSKGCGGGLPEVLEI 194
Query: 346 D-RYGVRILTSNPLYNQMNKENVDIGKVEEGFGLLLNNQDGRPRQVEVKA 394
+ G+RILTSNPLY K E+ GLL+ D P+QVEVKA
Sbjct: 195 NGNSGLRILTSNPLYQNAKK--------EKEIGLLVPEID-IPKQVEVKA 317
>AV765364
Length = 210
Score = 99.4 bits (246), Expect = 9e-22
Identities = 51/69 (73%), Positives = 54/69 (77%), Gaps = 1/69 (1%)
Frame = +3
Query: 128 ITMNGNHEIMNAEGDFRFATKNGVEEFKVWLEWFRQGNKMKNLCKGLEETVVDPLENVHV 187
ITMNGNHEIMN EGDFRFAT++GVEE+K WLEWFR GNKMK LC GLE DPLE V V
Sbjct: 3 ITMNGNHEIMNVEGDFRFATESGVEEYKSWLEWFRLGNKMKTLCNGLEPP-RDPLEGVTV 179
Query: 188 AFR-GVREE 195
R GVR E
Sbjct: 180 TPRGGVRSE 206
>AV411517
Length = 432
Score = 75.1 bits (183), Expect = 2e-14
Identities = 50/143 (34%), Positives = 71/143 (48%), Gaps = 5/143 (3%)
Frame = +3
Query: 109 ILYLLEKLKRQAAIHGGNFITMNGNHEIMNAEGDFRFATKNGVEEFKVWLEWFR-QGNKM 167
IL LL L++QA GG +NGNHE MN EGDFR+ G +E +LE+ G+
Sbjct: 6 ILSLLRSLEKQAKAKGGAVFQVNGNHETMNVEGDFRYVESGGFDECNDFLEYINDSGDDW 185
Query: 168 KNLCKGLEETVVDPLE--NVHVAFRGVRE--EFHDGFRARVAALRPNGPISKRFFTQNVT 223
+ G + E + ++ G + G AR RP+GP++ ++
Sbjct: 186 EETFTGWVDVSERWKEERKMSASYWGPWNLVKRQKGVIARSVLFRPSGPLACE-LARHAV 362
Query: 224 VLVVGDSIFVHGGLLKEHVDYGL 246
VL V D +F HGGLL HV YG+
Sbjct: 363 VLKVNDWVFCHGGLLPHHVAYGI 431
>CB827329
Length = 423
Score = 43.5 bits (101), Expect = 6e-05
Identities = 26/65 (40%), Positives = 37/65 (56%)
Frame = +1
Query: 20 SSFIDTFVDFSVSGGLFLPPPPSSPPPIPTRLPSPSRLIAIGDLHGDLKKSKEALSIAGL 79
S +D + S +G L P S PP P R+IA+GDLHGDL +++ AL +AG+
Sbjct: 217 SLLLDNNNNHSTAGSL-KPIVVSGNPPTFVSAPG-RRIIAVGDLHGDLHQARSALEMAGV 390
Query: 80 IDSSG 84
+ S G
Sbjct: 391 LSSDG 405
>TC16498 similar to GB|AAP37835.1|30725626|BT008476 At1g07010 {Arabidopsis
thaliana;}, partial (16%)
Length = 539
Score = 37.0 bits (84), Expect = 0.006
Identities = 22/49 (44%), Positives = 26/49 (52%)
Frame = +3
Query: 307 MGHTIQKEGINGVCENKAIRIDVGMSKGCGGGLPEVLEIDRYGVRILTS 355
+GHT Q G+N RIDVGMS G PEVLEI R++ S
Sbjct: 3 VGHTPQTIGVNCKYNCSIWRIDVGMSSGVLNSRPEVLEIIDDKARVIKS 149
>TC18015
Length = 460
Score = 33.1 bits (74), Expect = 0.082
Identities = 29/100 (29%), Positives = 42/100 (42%), Gaps = 1/100 (1%)
Frame = +1
Query: 48 PTRLPSPSRLIAIGDLHGDLKKSKEALS-IAGLIDSSGNYTGGSATVVQIGDVLDRGGDE 106
P P + IGD+HG + K + S + ID S T AT++ +GD DRG
Sbjct: 43 PMSEPESRVICCIGDIHGFITKLQNLWSNLEHSIDPSHFQT---ATIIFLGDYCDRGPHT 213
Query: 107 IKILYLLEKLKRQAAIHGGNFITMNGNHEIMNAEGDFRFA 146
++L L L + N H ++ DF FA
Sbjct: 214 REVLDFLIALPSRYP---------NQRHVFLSGNHDFAFA 306
>AV419982
Length = 415
Score = 32.7 bits (73), Expect = 0.11
Identities = 13/21 (61%), Positives = 15/21 (70%)
Frame = +2
Query: 40 PPSSPPPIPTRLPSPSRLIAI 60
PP SPPP P R PSPS L+ +
Sbjct: 251 PPRSPPPPPLRFPSPSLLLLL 313
>BP039310
Length = 584
Score = 32.7 bits (73), Expect = 0.11
Identities = 13/17 (76%), Positives = 13/17 (76%)
Frame = +2
Query: 35 LFLPPPPSSPPPIPTRL 51
LF PPPP PPPIP RL
Sbjct: 35 LFSPPPPWRPPPIPARL 85
>AV419295
Length = 383
Score = 32.7 bits (73), Expect = 0.11
Identities = 12/18 (66%), Positives = 15/18 (82%)
Frame = +1
Query: 38 PPPPSSPPPIPTRLPSPS 55
PPP SPPP PT+LP+P+
Sbjct: 157 PPPLPSPPPPPTKLPTPT 210
>BP043764
Length = 479
Score = 32.3 bits (72), Expect = 0.14
Identities = 14/28 (50%), Positives = 15/28 (53%)
Frame = +3
Query: 38 PPPPSSPPPIPTRLPSPSRLIAIGDLHG 65
PPPP PPP P LP P R I + G
Sbjct: 312 PPPPPPPPPPPPPLPPPERETEIAEDKG 395
>TC7812 UP|Q9LKJ6 (Q9LKJ6) Water-selective transport intrinsic membrane
protein 1, complete
Length = 1149
Score = 32.0 bits (71), Expect = 0.18
Identities = 13/18 (72%), Positives = 13/18 (72%)
Frame = +1
Query: 38 PPPPSSPPPIPTRLPSPS 55
PPP SSPPP PT PS S
Sbjct: 235 PPPASSPPPSPTHSPSSS 288
>TC13458 weakly similar to UP|Z282_HUMAN (Q9UDV7) Zinc finger protein 282
(HTLV-I U5RE binding protein 1) (HUB-1), partial (3%)
Length = 810
Score = 31.6 bits (70), Expect = 0.24
Identities = 12/17 (70%), Positives = 13/17 (75%)
Frame = +2
Query: 39 PPPSSPPPIPTRLPSPS 55
PPPS PPP P + PSPS
Sbjct: 302 PPPSPPPPSPKKPPSPS 352
>BP047567
Length = 480
Score = 31.2 bits (69), Expect = 0.31
Identities = 12/18 (66%), Positives = 14/18 (77%)
Frame = +2
Query: 40 PPSSPPPIPTRLPSPSRL 57
PP + PP+PT LPSPS L
Sbjct: 113 PPQTTPPLPTSLPSPSPL 166
>TC8542 similar to UP|RK1_PEA (P49208) 50S ribosomal protein L1,
chloroplast precursor (Fragment), partial (53%)
Length = 672
Score = 31.2 bits (69), Expect = 0.31
Identities = 14/28 (50%), Positives = 17/28 (60%)
Frame = +1
Query: 29 FSVSGGLFLPPPPSSPPPIPTRLPSPSR 56
F++S LP P +SPPP LPSP R
Sbjct: 166 FTLSFSSLLPTPSTSPPPTSGLLPSPPR 249
>BP039012
Length = 582
Score = 25.8 bits (55), Expect(2) = 0.32
Identities = 10/16 (62%), Positives = 11/16 (68%)
Frame = +1
Query: 30 SVSGGLFLPPPPSSPP 45
S SGG PPPP+ PP
Sbjct: 217 SHSGGSLSPPPPTPPP 264
Score = 23.9 bits (50), Expect(2) = 0.32
Identities = 11/28 (39%), Positives = 15/28 (53%), Gaps = 2/28 (7%)
Frame = +3
Query: 38 PPPPSSPPP--IPTRLPSPSRLIAIGDL 63
PPPP SP P I +P+ L + +L
Sbjct: 285 PPPPPSPSPSTITITIPNLQELFHVSEL 368
>AV410275
Length = 431
Score = 30.8 bits (68), Expect = 0.41
Identities = 13/31 (41%), Positives = 18/31 (57%)
Frame = +1
Query: 25 TFVDFSVSGGLFLPPPPSSPPPIPTRLPSPS 55
+F+ + L PPPP PPP P+ P+PS
Sbjct: 4 SFISLISTQSLPPPPPPPPPPPPPSWSPNPS 96
>BP064290
Length = 412
Score = 30.8 bits (68), Expect = 0.41
Identities = 12/18 (66%), Positives = 13/18 (71%)
Frame = +3
Query: 38 PPPPSSPPPIPTRLPSPS 55
PPP +PPP P LPSPS
Sbjct: 309 PPPSPAPPPPPRPLPSPS 362
>TC11625 weakly similar to UP|O81015 (O81015) At2g26920 protein, partial
(8%)
Length = 410
Score = 30.8 bits (68), Expect = 0.41
Identities = 27/102 (26%), Positives = 40/102 (38%), Gaps = 24/102 (23%)
Frame = -3
Query: 15 IPNFLSSFIDTFVDFSVSGGLF--------LPPPPSSPPPIPTRLPSPSRLIAIGDLHG- 65
+PN +S+ + + S+S L +PPPP PPP P P + L+++ + G
Sbjct: 390 LPNSVSASKNLSLSLSLSLSLHRRFSHSTVIPPPPPPPPPPP---PPSTTLLSLSETPGP 220
Query: 66 ---------------DLKKSKEALSIAGLIDSSGNYTGGSAT 92
D K KEA + SGN G T
Sbjct: 219 LST*EMSPASRSKSKDKKACKEAQKASAKPTGSGNAVAGIPT 94
>TC14009 similar to UP|HSF5_ARATH (Q9S7U5) Heat shock factor protein 5 (HSF
5) (Heat shock transcription factor 5) (HSTF 5), partial
(27%)
Length = 406
Score = 30.8 bits (68), Expect = 0.41
Identities = 18/57 (31%), Positives = 25/57 (43%)
Frame = +2
Query: 37 LPPPPSSPPPIPTRLPSPSRLIAIGDLHGDLKKSKEALSIAGLIDSSGNYTGGSATV 93
+PP PSSP P P PS S + L S +S A +S+ +G S +
Sbjct: 119 IPPTPSSPGPNPPATPSSSPTSPLSPSPSSLPISNTTISPASSANSTSMVSGKSIRI 289
Database: LJGI
Posted date: Jul 30, 2004 11:16 AM
Number of letters in database: 14,692,800
Number of sequences in database: 28,460
Lambda K H
0.319 0.141 0.425
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 7,202,094
Number of Sequences: 28460
Number of extensions: 124520
Number of successful extensions: 3534
Number of sequences better than 10.0: 191
Number of HSP's better than 10.0 without gapping: 2107
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 3046
length of query: 394
length of database: 4,897,600
effective HSP length: 92
effective length of query: 302
effective length of database: 2,279,280
effective search space: 688342560
effective search space used: 688342560
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 56 (26.2 bits)
Medicago: description of AC146752.2


