

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC144388.8 - phase: 0
(160 letters)
Database: LJGI
28,460 sequences; 14,692,800 total letters
Searching..................................................done
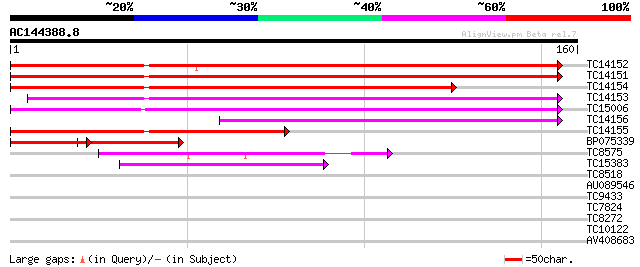
Score E
Sequences producing significant alignments: (bits) Value
TC14152 weakly similar to UP|AB18_PEA (Q06930) ABA-responsive pr... 130 8e-32
TC14151 similar to UP|P93333 (P93333) PR10-1 protein (Class 10 P... 130 8e-32
TC14154 similar to UP|P93333 (P93333) PR10-1 protein (Class 10 P... 110 1e-25
TC14153 weakly similar to UP|P93333 (P93333) PR10-1 protein (Cla... 109 3e-25
TC15006 weakly similar to UP|P93333 (P93333) PR10-1 protein (Cla... 106 2e-24
TC14156 similar to UP|P93333 (P93333) PR10-1 protein (Class 10 P... 81 7e-17
TC14155 weakly similar to UP|Q8W1M7 (Q8W1M7) Drought-induced pro... 80 2e-16
BP075339 40 2e-10
TC8575 weakly similar to UP|Q8L9L8 (Q8L9L8) Pollen allergen-like... 54 1e-08
TC15383 44 2e-05
TC8518 similar to UP|Q84MC7 (Q84MC7) At1g01360, partial (84%) 34 0.010
AU089546 31 0.089
TC9433 similar to UP|Q9M4Y9 (Q9M4Y9) AP2-related transcription f... 28 0.98
TC7824 similar to UP|O04919 (O04919) Lipoxygenase , partial (58%) 28 0.98
TC8272 similar to UP|AAQ72568 (AAQ72568) Ripening-related protei... 27 1.7
TC10122 26 2.9
AV408683 25 4.9
>TC14152 weakly similar to UP|AB18_PEA (Q06930) ABA-responsive protein
ABR18, partial (92%)
Length = 827
Score = 130 bits (328), Expect = 8e-32
Identities = 66/157 (42%), Positives = 101/157 (64%), Gaps = 1/157 (0%)
Frame = +1
Query: 1 MGVTTFTHEYSSTVAPSRMFTALIIDSRNLIPKLLPQFVKDVNIIEGDGGA-GSIEQVNF 59
M V TFT E +STVAP+++F AL++D ++PK++P F K V I+EG+G A G+++++
Sbjct: 91 MAVLTFTDETTSTVAPAKLFKALVLDVDTIVPKVIPVF-KSVEIVEGNGIAVGTVKKITI 267
Query: 60 NEGSPFKYLKQKIDVLDKENLICKYTMIEGDPLGDKLESIAYEVKFEATNDGGCLCKMAS 119
NEG KY+ KID +D+ N + Y++I GD L D E I++E K A DGG + K+
Sbjct: 268 NEGGEDKYVLHKIDAIDEANFVYNYSVIGGDGLPDAAEKISFESKLIAGPDGGSIAKLTV 447
Query: 120 SYKTIGDFDVKEEDVKEGRESTIGIYEVVESYLLENP 156
+ GD E ++KEG+E G+++ VE Y+L NP
Sbjct: 448 HFHLKGDAKPTEAELKEGKEKGDGLFKAVEGYVLANP 558
>TC14151 similar to UP|P93333 (P93333) PR10-1 protein (Class 10 PR protein),
complete
Length = 997
Score = 130 bits (328), Expect = 8e-32
Identities = 67/156 (42%), Positives = 99/156 (62%)
Frame = +3
Query: 1 MGVTTFTHEYSSTVAPSRMFTALIIDSRNLIPKLLPQFVKDVNIIEGDGGAGSIEQVNFN 60
MGV TF E +STVAP+R++ AL ID ++IPK+LP F K V I++G+G AG+I+++ F
Sbjct: 87 MGVFTFQDETTSTVAPARLYKALTIDGDSIIPKVLPGF-KSVEIVKGNGSAGTIKKITFE 263
Query: 61 EGSPFKYLKQKIDVLDKENLICKYTMIEGDPLGDKLESIAYEVKFEATNDGGCLCKMASS 120
E K L KI+ +D+ N Y++I G L D +E I++E K A DGG + K+ +
Sbjct: 264 EDGKTKDLLHKIESIDEANFGYSYSIIGGSDLPDTVEKISFEAKLVAGPDGGSIAKLTVN 443
Query: 121 YKTIGDFDVKEEDVKEGRESTIGIYEVVESYLLENP 156
Y T GD EE++K G+ G+++ VE Y+L NP
Sbjct: 444 YHTKGDASPSEEEIKAGKAKGDGLFKAVEGYVLANP 551
>TC14154 similar to UP|P93333 (P93333) PR10-1 protein (Class 10 PR protein),
partial (82%)
Length = 432
Score = 110 bits (275), Expect = 1e-25
Identities = 56/126 (44%), Positives = 83/126 (65%)
Frame = +2
Query: 1 MGVTTFTHEYSSTVAPSRMFTALIIDSRNLIPKLLPQFVKDVNIIEGDGGAGSIEQVNFN 60
MGV TF E +STVAP+R++ AL ID ++IPK++P F K V +EGDGG G+I+++ F
Sbjct: 47 MGVFTFQDETTSTVAPARLYKALTIDGDSIIPKVIPGF-KSVETVEGDGGVGTIKKLTFV 223
Query: 61 EGSPFKYLKQKIDVLDKENLICKYTMIEGDPLGDKLESIAYEVKFEATNDGGCLCKMASS 120
EG+ K L KI+ +D+ N Y+++ G L D +E I++E K A +DGG + K+ +
Sbjct: 224 EGAETKNLLHKIESIDEANFGYHYSIVGGSELPDTVEKISFEAKLVAGSDGGSIAKLTVN 403
Query: 121 YKTIGD 126
Y T GD
Sbjct: 404 YHTKGD 421
>TC14153 weakly similar to UP|P93333 (P93333) PR10-1 protein (Class 10 PR
protein), partial (97%)
Length = 754
Score = 109 bits (272), Expect = 3e-25
Identities = 56/151 (37%), Positives = 90/151 (59%)
Frame = +2
Query: 6 FTHEYSSTVAPSRMFTALIIDSRNLIPKLLPQFVKDVNIIEGDGGAGSIEQVNFNEGSPF 65
F E +STVAP+R++ AL ID +IPK++P F + V I+EG+GG G+I+++ F E
Sbjct: 113 FQDETTSTVAPARLYKALTIDGDTIIPKVIPGF-RTVEIVEGNGGPGTIKKLTFEEDGQT 289
Query: 66 KYLKQKIDVLDKENLICKYTMIEGDPLGDKLESIAYEVKFEATNDGGCLCKMASSYKTIG 125
K L KID +D+ NL Y+++ G L +E I++E ++GG + K +Y T G
Sbjct: 290 KELLHKIDSIDQANLKYNYSIVGGTDLPATVEKISFEAHLVVGSNGGSIAKHNINYHTKG 469
Query: 126 DFDVKEEDVKEGRESTIGIYEVVESYLLENP 156
+ EE+V G+ +++ +E Y+L NP
Sbjct: 470 NASFSEEEVMAGKAKGDALFKAIEGYVLANP 562
>TC15006 weakly similar to UP|P93333 (P93333) PR10-1 protein (Class 10 PR
protein), partial (75%)
Length = 1047
Score = 106 bits (264), Expect = 2e-24
Identities = 53/156 (33%), Positives = 91/156 (57%)
Frame = +1
Query: 1 MGVTTFTHEYSSTVAPSRMFTALIIDSRNLIPKLLPQFVKDVNIIEGDGGAGSIEQVNFN 60
MGV + + VAP+R+F A+ D N+ PK++ ++ V IEG G AG+I+++
Sbjct: 79 MGVVSDEFSTPAAVAPARLFKAMSTDFHNVFPKIVEP-IQSVEFIEGTGAAGTIKKMTVL 255
Query: 61 EGSPFKYLKQKIDVLDKENLICKYTMIEGDPLGDKLESIAYEVKFEATNDGGCLCKMASS 120
+G KY+ ++D +D++ L+ +++I G L D LE + ++ KF +GGC+ ++ +
Sbjct: 256 DGGESKYMLHRVDEVDEKELVYNFSIIGGTGLADPLEKVQFKSKFVEGPNGGCIREVQAQ 435
Query: 121 YKTIGDFDVKEEDVKEGRESTIGIYEVVESYLLENP 156
Y T GD + EE VK + GI ++ E +LL NP
Sbjct: 436 YFTKGDTTLSEETVKASQAKVNGIVKIAEGFLLANP 543
>TC14156 similar to UP|P93333 (P93333) PR10-1 protein (Class 10 PR protein),
partial (62%)
Length = 549
Score = 81.3 bits (199), Expect = 7e-17
Identities = 39/97 (40%), Positives = 58/97 (59%)
Frame = +3
Query: 60 NEGSPFKYLKQKIDVLDKENLICKYTMIEGDPLGDKLESIAYEVKFEATNDGGCLCKMAS 119
NEG KY+ KID +D+ N + Y++I GD L D E I++E K A +DGG + K+
Sbjct: 15 NEGGEDKYVLHKIDAIDEANFVYNYSVIGGDGLPDAAEKISFESKLIAGSDGGSIAKLTI 194
Query: 120 SYKTIGDFDVKEEDVKEGRESTIGIYEVVESYLLENP 156
+ GD E ++KEG+E G+++ VE Y+L NP
Sbjct: 195 HFHLKGDAKPTEAELKEGKEKGDGLFKAVEGYVLANP 305
>TC14155 weakly similar to UP|Q8W1M7 (Q8W1M7) Drought-induced protein
RPR-10, partial (50%)
Length = 324
Score = 80.1 bits (196), Expect = 2e-16
Identities = 37/79 (46%), Positives = 57/79 (71%)
Frame = +3
Query: 1 MGVTTFTHEYSSTVAPSRMFTALIIDSRNLIPKLLPQFVKDVNIIEGDGGAGSIEQVNFN 60
M V TFT E +STVAP+++ AL++D ++PK++P F K V I+EG+GGAG+++++ N
Sbjct: 90 MAVLTFTDETTSTVAPAKLSKALVLDVDTIVPKVIPVF-KSVAIVEGNGGAGTVKKITIN 266
Query: 61 EGSPFKYLKQKIDVLDKEN 79
EG KY+ KID +D+ N
Sbjct: 267 EGGEDKYVWHKIDAIDEAN 323
>BP075339
Length = 528
Score = 40.4 bits (93), Expect(2) = 2e-10
Identities = 18/30 (60%), Positives = 23/30 (76%)
Frame = +1
Query: 20 FTALIIDSRNLIPKLLPQFVKDVNIIEGDG 49
F LIIDS+NLIPKL + V DV +I+G+G
Sbjct: 427 FKGLIIDSKNLIPKL*AEIVNDVQVIQGEG 516
Score = 39.7 bits (91), Expect(2) = 2e-10
Identities = 17/23 (73%), Positives = 20/23 (86%)
Frame = +3
Query: 1 MGVTTFTHEYSSTVAPSRMFTAL 23
MGVTTFTHE+SS++APSRM L
Sbjct: 369 MGVTTFTHEFSSSIAPSRMVQGL 437
>TC8575 weakly similar to UP|Q8L9L8 (Q8L9L8) Pollen allergen-like protein,
partial (79%)
Length = 762
Score = 54.3 bits (129), Expect = 1e-08
Identities = 32/85 (37%), Positives = 43/85 (49%), Gaps = 2/85 (2%)
Frame = +3
Query: 26 DSRNLIPKLLPQFVKDVNIIEGDG-GAGSIEQVNFNEGSPF-KYLKQKIDVLDKENLICK 83
DS + PK P K + I+EGDG AGS+ + + EGSP K +KID D E
Sbjct: 105 DSTTIFPKAFPHDYKSIEILEGDGKAAGSVRHITYAEGSPLVKSSTEKIDAADDEKKTVS 284
Query: 84 YTMIEGDPLGDKLESIAYEVKFEAT 108
Y +I+G E + Y KF+ T
Sbjct: 285 YAVIDG-------ELLQYYKKFKGT 338
>TC15383
Length = 729
Score = 43.5 bits (101), Expect = 2e-05
Identities = 17/59 (28%), Positives = 35/59 (58%)
Frame = +3
Query: 32 PKLLPQFVKDVNIIEGDGGAGSIEQVNFNEGSPFKYLKQKIDVLDKENLICKYTMIEGD 90
P+ FVK ++I GDG G++ +V+ G P +++++LD+E + ++++ GD
Sbjct: 519 PQAYKHFVKSCHVIGGDGNVGTLREVHVISGLPAGRSTERLEILDEERHVLSFSVVGGD 695
>TC8518 similar to UP|Q84MC7 (Q84MC7) At1g01360, partial (84%)
Length = 665
Score = 34.3 bits (77), Expect = 0.010
Identities = 16/47 (34%), Positives = 26/47 (55%)
Frame = +2
Query: 44 IIEGDGGAGSIEQVNFNEGSPFKYLKQKIDVLDKENLICKYTMIEGD 90
I++GD G GS+ +VN G P ++++ LD E I ++ GD
Sbjct: 338 IMQGDLGIGSVREVNVKSGLPATTSTERLEQLDDEEHILGIRIVGGD 478
>AU089546
Length = 134
Score = 31.2 bits (69), Expect = 0.089
Identities = 13/26 (50%), Positives = 18/26 (69%)
Frame = +3
Query: 1 MGVTTFTHEYSSTVAPSRMFTALIID 26
M TFT E+++TV R+F ALI+D
Sbjct: 57 MDALTFTEEFANTVXAGRLFKALILD 134
>TC9433 similar to UP|Q9M4Y9 (Q9M4Y9) AP2-related transcription factor
(Ethylene responsive protein), partial (24%)
Length = 572
Score = 27.7 bits (60), Expect = 0.98
Identities = 12/28 (42%), Positives = 18/28 (63%)
Frame = +2
Query: 84 YTMIEGDPLGDKLESIAYEVKFEATNDG 111
+ + GDPL D+ E ++ E +FEAT G
Sbjct: 365 FFLFAGDPLSDEEEQLSDEDRFEATLTG 448
>TC7824 similar to UP|O04919 (O04919) Lipoxygenase , partial (58%)
Length = 1675
Score = 27.7 bits (60), Expect = 0.98
Identities = 15/43 (34%), Positives = 23/43 (52%)
Frame = -3
Query: 56 QVNFNEGSPFKYLKQKIDVLDKENLICKYTMIEGDPLGDKLES 98
Q N N S Y K ++ K + + YT+++GD +GD ES
Sbjct: 1595 QRN*NSLSHMNYFKATTTIIYKPSAL--YTLLDGDTIGDSPES 1473
>TC8272 similar to UP|AAQ72568 (AAQ72568) Ripening-related protein, partial
(64%)
Length = 757
Score = 26.9 bits (58), Expect = 1.7
Identities = 13/54 (24%), Positives = 28/54 (51%)
Frame = +3
Query: 69 KQKIDVLDKENLICKYTMIEGDPLGDKLESIAYEVKFEATNDGGCLCKMASSYK 122
+++I+ +D+EN KY++ +GD +G ++ ++ DG K Y+
Sbjct: 285 QERIESVDEENKTIKYSLFDGD-IGQHYKTFLLTLQVIENKDGHDTVKWTVEYE 443
>TC10122
Length = 544
Score = 26.2 bits (56), Expect = 2.9
Identities = 10/26 (38%), Positives = 16/26 (61%)
Frame = +3
Query: 32 PKLLPQFVKDVNIIEGDGGAGSIEQV 57
P+ FVK ++I GDG G++ +V
Sbjct: 453 PQAYKNFVKSCHVILGDGDVGTLREV 530
>AV408683
Length = 194
Score = 25.4 bits (54), Expect = 4.9
Identities = 16/53 (30%), Positives = 28/53 (52%)
Frame = +2
Query: 93 GDKLESIAYEVKFEATNDGGCLCKMASSYKTIGDFDVKEEDVKEGRESTIGIY 145
GD+L S Y+++F+ D +CK KT+ K++DV + R + + Y
Sbjct: 29 GDRLVSAPYKLEFQRDKDSVVVCK-----KTL-----KKDDVVKFRSAVMKDY 157
Database: LJGI
Posted date: Jul 30, 2004 11:16 AM
Number of letters in database: 14,692,800
Number of sequences in database: 28,460
Lambda K H
0.315 0.137 0.386
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 2,159,447
Number of Sequences: 28460
Number of extensions: 23223
Number of successful extensions: 71
Number of sequences better than 10.0: 34
Number of HSP's better than 10.0 without gapping: 66
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 66
length of query: 160
length of database: 4,897,600
effective HSP length: 83
effective length of query: 77
effective length of database: 2,535,420
effective search space: 195227340
effective search space used: 195227340
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (22.0 bits)
S2: 51 (24.3 bits)
Medicago: description of AC144388.8


