

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC140104.12 - phase: 0
(279 letters)
Database: LJGI
28,460 sequences; 14,692,800 total letters
Searching..................................................done
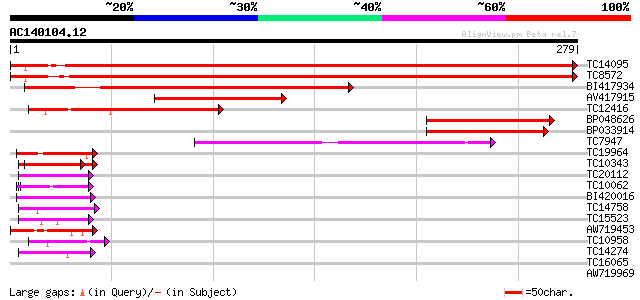
Score E
Sequences producing significant alignments: (bits) Value
TC14095 homologue to UP|RS2_ARATH (P49688) 40S ribosomal protein... 525 e-150
TC8572 similar to UP|Q9SCM3 (Q9SCM3) 40S ribosomal protein S2 ho... 517 e-148
BI417934 204 2e-53
AV417915 131 1e-31
TC12416 similar to GB|AAH56066.1|33417132|BC056066 sop-prov prot... 113 3e-26
BP048626 112 5e-26
BP033914 91 2e-19
TC7947 similar to UP|Q9ST69 (Q9ST69) Ribosomal protein S5, parti... 76 7e-15
TC19964 similar to UP|Q40363 (Q40363) NuM1 protein, partial (11%) 55 2e-08
TC10343 similar to UP|Q9SZZ1 (Q9SZZ1) Fibrillarin-like protein (... 45 1e-05
TC20112 weakly similar to UP|Q9SBM1 (Q9SBM1) Hydroxyproline-rich... 43 7e-05
TC10062 similar to BAD08700 (BAD08700) Cold shock domain protein... 42 1e-04
BI420016 42 2e-04
TC14758 similar to UP|O24601 (O24601) Glycine-rich RNA binding p... 42 2e-04
TC15523 similar to UP|GRPA_MAIZE (P10979) Glycine-rich RNA-bindi... 41 2e-04
AW719453 41 2e-04
TC10958 similar to PIR|T47938|T47938 RING finger protein - Arabi... 40 3e-04
TC14274 weakly similar to GB|AAF79559.1|8778551|AC022464 F22G5.3... 40 4e-04
TC16065 weakly similar to UP|Q9SBM1 (Q9SBM1) Hydroxyproline-rich... 38 0.002
AW719969 38 0.002
>TC14095 homologue to UP|RS2_ARATH (P49688) 40S ribosomal protein S2,
partial (88%)
Length = 1134
Score = 525 bits (1353), Expect = e-150
Identities = 265/280 (94%), Positives = 270/280 (95%), Gaps = 1/280 (0%)
Frame = +3
Query: 1 MAERGG-DRGGFGSGFGGRGGDRGGRGGRGDRGRGGRRRAGGRREEEEKWVPVTKLGRLV 59
MAERGG DRGGFG GFGGRG DRG RGDRGRGGRRR GGRR+EEEKWVPVTKLGRLV
Sbjct: 54 MAERGGGDRGGFGRGFGGRG-DRG----RGDRGRGGRRRTGGRRDEEEKWVPVTKLGRLV 218
Query: 60 KEGKIRSLEQIYLHSLPIKEHQIIDTLVGPTLKDEVMKIMPVQKQTRAGQRTRFKAFVVV 119
++GKIRSLEQIYLHSLPIKEHQIID LVGPTLKDEVMKIMPVQKQTRAGQRTRFKAFVVV
Sbjct: 219 RDGKIRSLEQIYLHSLPIKEHQIIDMLVGPTLKDEVMKIMPVQKQTRAGQRTRFKAFVVV 398
Query: 120 GDNNGHVGLGVKCSKEVATAIRGAIILAKLSVIPVRRGYWGNKIGKPHTVPCKVTGKCGS 179
GDNNGHVGLGVKCSKEVATAIRG IILAKLSVIPVRRGYWGNKIGKPHTVPCKVTGKCGS
Sbjct: 399 GDNNGHVGLGVKCSKEVATAIRGGIILAKLSVIPVRRGYWGNKIGKPHTVPCKVTGKCGS 578
Query: 180 VTVRMVPAPRGSGIVAARVPKKVLQFAGIDDVFTSSRGSTKTLGNFVKATFDCLMKTYGF 239
VTVRMVPAPRGSGIVAARVPKKVLQFAGIDDVFTSSRGSTKTLGNFVKATFDCL+KTYGF
Sbjct: 579 VTVRMVPAPRGSGIVAARVPKKVLQFAGIDDVFTSSRGSTKTLGNFVKATFDCLLKTYGF 758
Query: 240 LTPEFWKETRFSKSPFQEYTDLLAKPTAKALILEEERVEA 279
LTPEFWKETRF+KSPFQEYTDLLAKPTAKALILEEERVEA
Sbjct: 759 LTPEFWKETRFTKSPFQEYTDLLAKPTAKALILEEERVEA 878
>TC8572 similar to UP|Q9SCM3 (Q9SCM3) 40S ribosomal protein S2 homolog
(At3g57490), partial (91%)
Length = 1076
Score = 517 bits (1332), Expect = e-148
Identities = 262/285 (91%), Positives = 268/285 (93%), Gaps = 6/285 (2%)
Frame = +2
Query: 1 MAERGG------DRGGFGSGFGGRGGDRGGRGGRGDRGRGGRRRAGGRREEEEKWVPVTK 54
MAERGG DRGGFG GFGGRG RGG DRGRGGRRR GRR+EEEKWVPVTK
Sbjct: 65 MAERGGGERGGGDRGGFGRGFGGRG-----RGG--DRGRGGRRRGAGRRDEEEKWVPVTK 223
Query: 55 LGRLVKEGKIRSLEQIYLHSLPIKEHQIIDTLVGPTLKDEVMKIMPVQKQTRAGQRTRFK 114
LGRLV++GKIRSLEQIYLHSLPIKEHQIIDTLVGPTLKDEVMKIMPVQKQTRAGQRTRFK
Sbjct: 224 LGRLVRDGKIRSLEQIYLHSLPIKEHQIIDTLVGPTLKDEVMKIMPVQKQTRAGQRTRFK 403
Query: 115 AFVVVGDNNGHVGLGVKCSKEVATAIRGAIILAKLSVIPVRRGYWGNKIGKPHTVPCKVT 174
AFVVVGDNNGHVGLGVKCSKEVATAIRG IILAKLSVIPVRRGYWGNKIGKPHTVPCKVT
Sbjct: 404 AFVVVGDNNGHVGLGVKCSKEVATAIRGGIILAKLSVIPVRRGYWGNKIGKPHTVPCKVT 583
Query: 175 GKCGSVTVRMVPAPRGSGIVAARVPKKVLQFAGIDDVFTSSRGSTKTLGNFVKATFDCLM 234
GKCGSVTVRMVPAPRGSGIVAARVPKKVLQFAGIDDVFTSSRGSTKTLGNFVKATFDCL+
Sbjct: 584 GKCGSVTVRMVPAPRGSGIVAARVPKKVLQFAGIDDVFTSSRGSTKTLGNFVKATFDCLL 763
Query: 235 KTYGFLTPEFWKETRFSKSPFQEYTDLLAKPTAKALILEEERVEA 279
KTYGFLTPEFWKETRF+KSPFQEYTDLLAKPTAKALILEEERV+A
Sbjct: 764 KTYGFLTPEFWKETRFTKSPFQEYTDLLAKPTAKALILEEERVDA 898
>BI417934
Length = 502
Score = 204 bits (518), Expect = 2e-53
Identities = 102/162 (62%), Positives = 123/162 (74%)
Frame = +1
Query: 8 RGGFGSGFGGRGGDRGGRGGRGDRGRGGRRRAGGRREEEEKWVPVTKLGRLVKEGKIRSL 67
RGGFG G R R RG++G E+E+W PVTKLGRLVK+GKI+S+
Sbjct: 52 RGGFGRRREGGDAPRAPRPRRGEKG------------EKEEWTPVTKLGRLVKDGKIKSV 195
Query: 68 EQIYLHSLPIKEHQIIDTLVGPTLKDEVMKIMPVQKQTRAGQRTRFKAFVVVGDNNGHVG 127
E+I+L S+PIKE+QI+D L+G L+DEVMKIMPVQKQT AGQRTRFK +V VGD NGH+G
Sbjct: 196 EEIFLFSMPIKEYQIVDFLLGSALRDEVMKIMPVQKQTTAGQRTRFKCYVAVGDENGHIG 375
Query: 128 LGVKCSKEVATAIRGAIILAKLSVIPVRRGYWGNKIGKPHTV 169
LGVK +KEVATAIRG II AK+++IPVRRGYWG G+PHTV
Sbjct: 376 LGVKAAKEVATAIRGGIINAKMNLIPVRRGYWGLNAGEPHTV 501
>AV417915
Length = 303
Score = 131 bits (329), Expect = 1e-31
Identities = 64/65 (98%), Positives = 64/65 (98%)
Frame = +2
Query: 72 LHSLPIKEHQIIDTLVGPTLKDEVMKIMPVQKQTRAGQRTRFKAFVVVGDNNGHVGLGVK 131
LHSLPIKEHQIID LVGPTLKDEVMKIMPVQKQTRAGQRTRFKAFVVVGDNNGHVGLGVK
Sbjct: 107 LHSLPIKEHQIIDMLVGPTLKDEVMKIMPVQKQTRAGQRTRFKAFVVVGDNNGHVGLGVK 286
Query: 132 CSKEV 136
CSKEV
Sbjct: 287 CSKEV 301
>TC12416 similar to GB|AAH56066.1|33417132|BC056066 sop-prov protein
{Xenopus laevis;} , partial (32%)
Length = 416
Score = 113 bits (283), Expect = 3e-26
Identities = 65/99 (65%), Positives = 73/99 (73%), Gaps = 3/99 (3%)
Frame = +3
Query: 10 GFGSGFG-GRGGDRGGRGGRGDRGRGGRRRAGGRREEEEK--WVPVTKLGRLVKEGKIRS 66
GFG GFG GRG RG GRG RGRG R GRR + +K WVPVTKLGRLVK G I+S
Sbjct: 123 GFGDGFGRGRGEGRGRGEGRG-RGRGRGRGRRGRRGDGDKKEWVPVTKLGRLVKAGMIKS 299
Query: 67 LEQIYLHSLPIKEHQIIDTLVGPTLKDEVMKIMPVQKQT 105
LE+IY+ SL IKE +IID +G +LKDEVMKI PVQKQT
Sbjct: 300 LEEIYVFSLAIKEFEIIDHFLGDSLKDEVMKISPVQKQT 416
>BP048626
Length = 357
Score = 112 bits (281), Expect = 5e-26
Identities = 53/63 (84%), Positives = 58/63 (91%)
Frame = -3
Query: 206 AGIDDVFTSSRGSTKTLGNFVKATFDCLMKTYGFLTPEFWKETRFSKSPFQEYTDLLAKP 265
AGIDDVFTSSRGS +T GNFVKATFDCL+KTYGFLTP FWKETRF++SPF EYTDLLA P
Sbjct: 355 AGIDDVFTSSRGSPQTPGNFVKATFDCLLKTYGFLTPGFWKETRFTQSPFPEYTDLLA*P 176
Query: 266 TAK 268
TA+
Sbjct: 175 TAQ 167
>BP033914
Length = 350
Score = 90.9 bits (224), Expect = 2e-19
Identities = 38/60 (63%), Positives = 52/60 (86%)
Frame = -2
Query: 206 AGIDDVFTSSRGSTKTLGNFVKATFDCLMKTYGFLTPEFWKETRFSKSPFQEYTDLLAKP 265
AGI+D +T SRGST+TLGNFV+A F L +TYGFLTPE W+E++++++PFQE+TD LA+P
Sbjct: 349 AGIEDCYTCSRGSTQTLGNFVRAPFHALSRTYGFLTPELWRESKYTQAPFQEFTDFLAQP 170
>TC7947 similar to UP|Q9ST69 (Q9ST69) Ribosomal protein S5, partial (81%)
Length = 1294
Score = 75.9 bits (185), Expect = 7e-15
Identities = 44/148 (29%), Positives = 80/148 (53%)
Frame = +1
Query: 92 KDEVMKIMPVQKQTRAGQRTRFKAFVVVGDNNGHVGLGVKCSKEVATAIRGAIILAKLSV 151
++ V+++ + K + G++ +F+A VVVGD G+VG+GV +KEV A++ + + A+ ++
Sbjct: 553 EERVVQVRRITKVVKGGKQLKFRAVVVVGDKKGNVGVGVGKAKEVVAAVQKSAVNARRNI 732
Query: 152 IPVRRGYWGNKIGKPHTVPCKVTGKCGSVTVRMVPAPRGSGIVAARVPKKVLQFAGIDDV 211
+ V + K T P + G G+ V + PA G+G++A + VL+ AG+++
Sbjct: 733 VSV-------PMTKYLTFPHRSDGDYGAAKVMLRPASPGTGVIAGGAVRIVLEMAGVENA 891
Query: 212 FTSSRGSTKTLGNFVKATFDCLMKTYGF 239
GS L N +AT + K F
Sbjct: 892 LGKQLGSNNALNN-ARATVVAVQKMRQF 972
Score = 28.1 bits (61), Expect = 1.7
Identities = 22/58 (37%), Positives = 26/58 (43%), Gaps = 3/58 (5%)
Frame = -2
Query: 6 GDRGGFGSGFGGRGGDRGGRGGRGDRGRGGRRRAGGRR---EEEEKWVPVTKLGRLVK 60
G G G G GG G+RG G R R GRR ++E WV LG LV+
Sbjct: 189 GG*GEEGEG*GGAEGERGESSGGYYRRCSSH*RKEGRRLILKDEVGWVGSLCLGDLVE 16
>TC19964 similar to UP|Q40363 (Q40363) NuM1 protein, partial (11%)
Length = 562
Score = 54.7 bits (130), Expect = 2e-08
Identities = 29/43 (67%), Positives = 31/43 (71%), Gaps = 3/43 (6%)
Frame = +2
Query: 4 RGGDRGGFGSGFGGRGGDRGGRGGRGDRGRGGR---RRAGGRR 43
RGG RG FGSG GRGGDRGGRGG G GRGGR RA G++
Sbjct: 203 RGGGRGRFGSG--GRGGDRGGRGGSGGGGRGGRGTPLRAAGKK 325
Score = 35.4 bits (80), Expect = 0.011
Identities = 24/40 (60%), Positives = 24/40 (60%), Gaps = 2/40 (5%)
Frame = +2
Query: 5 GGDRGG-FGSGFGGRGGDRGGRGGR-GDRGRGGRRRAGGR 42
GG RGG F SG GG G R G GGR GDRG G GGR
Sbjct: 170 GGGRGGRFDSGRGG-GRGRFGSGGRGGDRGGRGGSGGGGR 286
Score = 35.0 bits (79), Expect = 0.014
Identities = 22/40 (55%), Positives = 22/40 (55%), Gaps = 2/40 (5%)
Frame = +2
Query: 6 GDRGGFGSGFGGRGG--DRGGRGGRGDRGRGGRRRAGGRR 43
G R G G GGRGG D G GGRG G GGR GG R
Sbjct: 143 GGRSGGGRSGGGRGGRFDSGRGGGRGRFGSGGR---GGDR 253
>TC10343 similar to UP|Q9SZZ1 (Q9SZZ1) Fibrillarin-like protein (Fibrillarin
2) (AtFib2), partial (85%)
Length = 1116
Score = 45.4 bits (106), Expect = 1e-05
Identities = 22/39 (56%), Positives = 24/39 (61%)
Frame = +1
Query: 5 GGDRGGFGSGFGGRGGDRGGRGGRGDRGRGGRRRAGGRR 43
GG GG G+ F RGG RGG GGRG RG G R GG +
Sbjct: 112 GGRGGGRGTPFKARGGGRGGGGGRGGRGGGRGGRGGGMK 228
Score = 40.0 bits (92), Expect = 4e-04
Identities = 19/30 (63%), Positives = 20/30 (66%)
Frame = +1
Query: 8 RGGFGSGFGGRGGDRGGRGGRGDRGRGGRR 37
RGG G GGRGG GGRGGRG +GG R
Sbjct: 151 RGGGRGGGGGRGGRGGGRGGRGGGMKGGSR 240
Score = 35.4 bits (80), Expect = 0.011
Identities = 25/45 (55%), Positives = 26/45 (57%), Gaps = 9/45 (20%)
Frame = +1
Query: 8 RGGFGSGFGGRG------GDRGGR-GGRGD--RGRGGRRRAGGRR 43
RGG G GF GRG GD GGR GGRG + RGG R GG R
Sbjct: 52 RGG-GGGFRGRGDRGRGRGDGGGRGGGRGTPFKARGGGRGGGGGR 183
Score = 31.2 bits (69), Expect = 0.20
Identities = 20/42 (47%), Positives = 20/42 (47%), Gaps = 6/42 (14%)
Frame = +1
Query: 8 RGGFGSGFGGRGGDRGGRG------GRGDRGRGGRRRAGGRR 43
RG G G G GG GGRG G G G GGR GG R
Sbjct: 79 RGDRGRGRGDGGGRGGGRGTPFKARGGGRGGGGGRGGRGGGR 204
Score = 30.4 bits (67), Expect = 0.35
Identities = 15/23 (65%), Positives = 16/23 (69%)
Frame = +1
Query: 5 GGDRGGFGSGFGGRGGDRGGRGG 27
GG RGG G G GGRGG G +GG
Sbjct: 172 GGGRGGRGGGRGGRGG--GMKGG 234
>TC20112 weakly similar to UP|Q9SBM1 (Q9SBM1) Hydroxyproline-rich
glycoprotein DZ-HRGP precursor, partial (11%)
Length = 523
Score = 42.7 bits (99), Expect = 7e-05
Identities = 22/37 (59%), Positives = 22/37 (59%)
Frame = +2
Query: 5 GGDRGGFGSGFGGRGGDRGGRGGRGDRGRGGRRRAGG 41
GG G G GFGGRGGDR GR G GGR R GG
Sbjct: 344 GGFSVGRGGGFGGRGGDRKYDTGRYSGGGGGRGRGGG 454
Score = 35.0 bits (79), Expect = 0.014
Identities = 25/47 (53%), Positives = 26/47 (55%), Gaps = 9/47 (19%)
Frame = +2
Query: 4 RGGDR----GGFGSGFGGRGGDRGG----RGGRGDRGRG-GRRRAGG 41
RGGDR G + G GGRG RGG GGRG RG G G R GG
Sbjct: 383 RGGDRKYDTGRYSGGGGGRG--RGGGSIRGGGRGGRGGGFGGRHGGG 517
>TC10062 similar to BAD08700 (BAD08700) Cold shock domain protein 2, partial
(66%)
Length = 481
Score = 42.0 bits (97), Expect = 1e-04
Identities = 22/38 (57%), Positives = 23/38 (59%)
Frame = +3
Query: 4 RGGDRGGFGSGFGGRGGDRGGRGGRGDRGRGGRRRAGG 41
RGG G G G+GG GG GG GG G GRGG R GG
Sbjct: 363 RGGGGYGGGGGYGGSGG-YGGGGGYGGGGRGGGRGGGG 473
Score = 40.8 bits (94), Expect = 3e-04
Identities = 21/38 (55%), Positives = 21/38 (55%)
Frame = +3
Query: 4 RGGDRGGFGSGFGGRGGDRGGRGGRGDRGRGGRRRAGG 41
RGG GG G G GG GG G GG G GRGG GG
Sbjct: 276 RGGGAGGGGRGGGGYGGGGYGGGGYGGGGRGGGGYGGG 389
Score = 40.0 bits (92), Expect = 4e-04
Identities = 21/37 (56%), Positives = 21/37 (56%)
Frame = +3
Query: 5 GGDRGGFGSGFGGRGGDRGGRGGRGDRGRGGRRRAGG 41
GG RGG G G GG GG G GGRG G GG GG
Sbjct: 294 GGGRGGGGYGGGGYGGGGYGGGGRGGGGYGGGGGYGG 404
Score = 39.3 bits (90), Expect = 7e-04
Identities = 20/36 (55%), Positives = 21/36 (57%)
Frame = +3
Query: 6 GDRGGFGSGFGGRGGDRGGRGGRGDRGRGGRRRAGG 41
G R G G+G GGRGG G GG G G GG R GG
Sbjct: 267 GTRRGGGAGGGGRGGGGYGGGGYGGGGYGGGGRGGG 374
Score = 34.7 bits (78), Expect = 0.018
Identities = 16/23 (69%), Positives = 16/23 (69%)
Frame = +3
Query: 5 GGDRGGFGSGFGGRGGDRGGRGG 27
GG GG G G GGRGG RGG GG
Sbjct: 408 GGYGGGGGYGGGGRGGGRGGGGG 476
>BI420016
Length = 559
Score = 41.6 bits (96), Expect = 2e-04
Identities = 22/39 (56%), Positives = 23/39 (58%)
Frame = -1
Query: 4 RGGDRGGFGSGFGGRGGDRGGRGGRGDRGRGGRRRAGGR 42
RGG GG G G GG RG RGG G + RG RRA GR
Sbjct: 499 RGGGGGGRGRGRREEGGRRGQRGGGGGQRRGRCRRASGR 383
Score = 34.7 bits (78), Expect = 0.018
Identities = 26/68 (38%), Positives = 26/68 (38%), Gaps = 21/68 (30%)
Frame = -1
Query: 3 ERGGDRGGFGSGFG----------GRGGDRGG-----------RGGRGDRGRGGRRRAGG 41
E GG RG G G G GRGG G RG RG R RG R R G
Sbjct: 460 EEGGRRGQRGGGGGQRRGRCRRASGRGGRIGSRRGRRRRRSRRRGRRGGRERGVRSRRGR 281
Query: 42 RREEEEKW 49
R E W
Sbjct: 280 RGRRSESW 257
Score = 26.6 bits (57), Expect = 5.0
Identities = 16/27 (59%), Positives = 16/27 (59%), Gaps = 4/27 (14%)
Frame = -1
Query: 21 DRGG----RGGRGDRGRGGRRRAGGRR 43
D GG GG G RGRG RR GGRR
Sbjct: 520 DHGGCCRRGGGGGGRGRG-RREEGGRR 443
>TC14758 similar to UP|O24601 (O24601) Glycine-rich RNA binding protein 2,
partial (93%)
Length = 725
Score = 41.6 bits (96), Expect = 2e-04
Identities = 24/49 (48%), Positives = 25/49 (50%), Gaps = 9/49 (18%)
Frame = +1
Query: 5 GGDRGGFG---------SGFGGRGGDRGGRGGRGDRGRGGRRRAGGRRE 44
GG RGG G G+GG GG RG GG G G GG GGRRE
Sbjct: 337 GGGRGGGGYGGGGGRREGGYGGGGGSRGYGGGGGGYGGGGGGGYGGRRE 483
Score = 34.3 bits (77), Expect = 0.024
Identities = 19/36 (52%), Positives = 20/36 (54%)
Frame = +1
Query: 8 RGGFGSGFGGRGGDRGGRGGRGDRGRGGRRRAGGRR 43
RGG G G G GG GG GGR + G GG GG R
Sbjct: 319 RGGGGGGGGRGGGGYGGGGGRREGGYGG---GGGSR 417
Score = 32.3 bits (72), Expect = 0.092
Identities = 15/23 (65%), Positives = 15/23 (65%)
Frame = +1
Query: 22 RGGRGGRGDRGRGGRRRAGGRRE 44
RGG GG G RG GG GGRRE
Sbjct: 319 RGGGGGGGGRGGGGYGGGGGRRE 387
>TC15523 similar to UP|GRPA_MAIZE (P10979) Glycine-rich RNA-binding,
abscisic acid-inducible protein, complete
Length = 972
Score = 41.2 bits (95), Expect = 2e-04
Identities = 23/48 (47%), Positives = 25/48 (51%), Gaps = 11/48 (22%)
Frame = +2
Query: 5 GGDRGGFGSG-----FGGRGGDR------GGRGGRGDRGRGGRRRAGG 41
GG GG+G G +GG GG R GG GG G G GGRR GG
Sbjct: 608 GGGGGGYGGGRREGGYGGGGGSRYSGGGGGGYGGSGGGGYGGRREGGG 751
Score = 28.9 bits (63), Expect = 1.0
Identities = 15/28 (53%), Positives = 16/28 (56%)
Frame = +2
Query: 14 GFGGRGGDRGGRGGRGDRGRGGRRRAGG 41
G GG GG GGR G G GG R +GG
Sbjct: 605 GGGGGGGYGGGRREGGYGGGGGSRYSGG 688
>AW719453
Length = 546
Score = 41.2 bits (95), Expect = 2e-04
Identities = 28/46 (60%), Positives = 29/46 (62%), Gaps = 3/46 (6%)
Frame = -2
Query: 1 MAERGGDRGGFGSGFGGRGGDRGGRGGRG--DRGRG-GRRRAGGRR 43
+ ER RGG G GFGG GG R GRGGRG GRG G R GGRR
Sbjct: 440 LQERDQSRGGRG-GFGG-GGSRFGRGGRGFSSGGRGYGGRGGGGRR 309
>TC10958 similar to PIR|T47938|T47938 RING finger protein - Arabidopsis
thaliana {Arabidopsis thaliana;}, partial (38%)
Length = 779
Score = 40.4 bits (93), Expect = 3e-04
Identities = 25/50 (50%), Positives = 26/50 (52%), Gaps = 10/50 (20%)
Frame = -3
Query: 10 GFGSGFGG----------RGGDRGGRGGRGDRGRGGRRRAGGRREEEEKW 49
G+GSG GG RGG RGG GGRG R R G RR GGR W
Sbjct: 267 GWGSGGGGPVRRRRGRRCRGGRRGGGGGRGRRRRSGERR-GGREGGGTVW 121
>TC14274 weakly similar to GB|AAF79559.1|8778551|AC022464 F22G5.35
{Arabidopsis thaliana;} , partial (16%)
Length = 1208
Score = 40.0 bits (92), Expect = 4e-04
Identities = 23/44 (52%), Positives = 24/44 (54%), Gaps = 6/44 (13%)
Frame = -1
Query: 5 GGDRGGFGSGFGGRGGDRGGRGG------RGDRGRGGRRRAGGR 42
GGD+GG G RGG R G GG R GRGGRR GGR
Sbjct: 461 GGDQGGRA*GGSCRGGGRRGGGG*ICGCWRNHGGRGGRRGGGGR 330
Score = 33.5 bits (75), Expect = 0.041
Identities = 20/37 (54%), Positives = 21/37 (56%)
Frame = -1
Query: 4 RGGDRGGFGSGFGGRGGDRGGRGGRGDRGRGGRRRAG 40
RGG R G G G + GGRGGR RG GGR R G
Sbjct: 422 RGGGRRGGGG*ICGCWRNHGGRGGR--RGGGGRWRRG 318
Score = 32.0 bits (71), Expect = 0.12
Identities = 17/34 (50%), Positives = 20/34 (58%), Gaps = 2/34 (5%)
Frame = -1
Query: 10 GFGSGFGGRG--GDRGGRGGRGDRGRGGRRRAGG 41
G+ G+G RG GD+GGR G GGRR GG
Sbjct: 494 GWSLGWGNRGVGGDQGGRA*GGSCRGGGRRGGGG 393
>TC16065 weakly similar to UP|Q9SBM1 (Q9SBM1) Hydroxyproline-rich
glycoprotein DZ-HRGP precursor, partial (20%)
Length = 608
Score = 37.7 bits (86), Expect = 0.002
Identities = 23/52 (44%), Positives = 26/52 (49%), Gaps = 13/52 (25%)
Frame = -1
Query: 4 RGGDRGGFGSGFGGRGGDRGGRGGRG-------------DRGRGGRRRAGGR 42
+GG GG+G G GG G GGRGG G RGRGG + GGR
Sbjct: 320 QGGRGGGYGGG-GGPGYGGGGRGGHGVPQQQYGGPPEYQGRGRGGPSQQGGR 168
Score = 33.9 bits (76), Expect = 0.032
Identities = 20/37 (54%), Positives = 21/37 (56%), Gaps = 1/37 (2%)
Frame = -1
Query: 6 GDRGGFGSGFGGRGGDRGGRGGRG-DRGRGGRRRAGG 41
G RGG+ G GG GG GGRGG G GRG GG
Sbjct: 176 GGRGGYSGGGGGYGGG-GGRGGGGMGSGRGVSPSHGG 69
Score = 33.5 bits (75), Expect = 0.041
Identities = 20/52 (38%), Positives = 25/52 (47%)
Frame = -1
Query: 6 GDRGGFGSGFGGRGGDRGGRGGRGDRGRGGRRRAGGRREEEEKWVPVTKLGR 57
G +GG G G +GG GG GG G G GG R G +++ P GR
Sbjct: 356 GPQGGRGYGGPPQGGRGGGYGGGGGPGYGGGGRGGHGVPQQQYGGPPEYQGR 201
Score = 32.0 bits (71), Expect = 0.12
Identities = 21/40 (52%), Positives = 22/40 (54%), Gaps = 1/40 (2%)
Frame = -1
Query: 4 RGGDRGGFGSGFGGRGGDRGGRGGRGDRG-RGGRRRAGGR 42
+G RGG S GGRGG GG GG G G RGG GR
Sbjct: 209 QGRGRGG-PSQQGGRGGYSGGGGGYGGGGGRGGGGMGSGR 93
Score = 27.3 bits (59), Expect = 2.9
Identities = 19/42 (45%), Positives = 20/42 (47%)
Frame = -1
Query: 2 AERGGDRGGFGSGFGGRGGDRGGRGGRGDRGRGGRRRAGGRR 43
A GG G G G G +GGRGG G G GG GG R
Sbjct: 380 AAPGGGGPGPQGGRGYGGPPQGGRGG-GYGGGGGPGYGGGGR 258
>AW719969
Length = 263
Score = 37.7 bits (86), Expect = 0.002
Identities = 23/43 (53%), Positives = 25/43 (57%)
Frame = -2
Query: 1 MAERGGDRGGFGSGFGGRGGDRGGRGGRGDRGRGGRRRAGGRR 43
+ ERGG GFGS RGG+ G GG G R RG RR GG R
Sbjct: 199 ICERGGSESGFGSRVRRRGGEVGVCGG*G-RIRGLRRGGGGWR 74
Database: LJGI
Posted date: Jul 30, 2004 11:16 AM
Number of letters in database: 14,692,800
Number of sequences in database: 28,460
Lambda K H
0.319 0.140 0.418
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 4,338,126
Number of Sequences: 28460
Number of extensions: 60293
Number of successful extensions: 1625
Number of sequences better than 10.0: 256
Number of HSP's better than 10.0 without gapping: 1039
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1387
length of query: 279
length of database: 4,897,600
effective HSP length: 89
effective length of query: 190
effective length of database: 2,364,660
effective search space: 449285400
effective search space used: 449285400
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 55 (25.8 bits)
Medicago: description of AC140104.12


