

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC138526.4 + phase: 0
(308 letters)
Database: LJGI
28,460 sequences; 14,692,800 total letters
Searching..................................................done
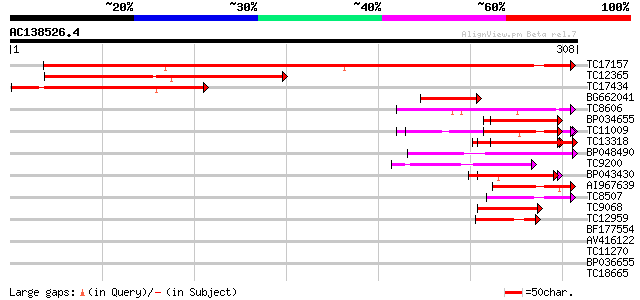
Score E
Sequences producing significant alignments: (bits) Value
TC17157 similar to UP|CAD27462 (CAD27462) Nucleosome assembly pr... 358 e-100
TC12365 similar to UP|Q39892 (Q39892) Nucleosome assembly protei... 185 1e-47
TC17434 similar to UP|Q94K07 (Q94K07) Nucleosome assembly protei... 127 3e-30
BG662041 61 3e-10
TC8606 similar to UP|AAQ82688 (AAQ82688) Epa4p, partial (7%) 55 1e-08
BP034655 51 3e-07
TC11009 similar to GB|AAP21377.1|30102918|BT006569 At1g47970 {Ar... 49 8e-07
TC13318 similar to UP|CAA20314 (CAA20314) SPBC30B4.01c protein (... 47 4e-06
BP048490 42 1e-04
TC9200 similar to UP|Q8LLE6 (Q8LLE6) RNA-binding protein AKIP1, ... 42 1e-04
BP043430 40 5e-04
AI967639 40 5e-04
TC8507 similar to UP|Q8LJS2 (Q8LJS2) Nucleolar histone deacetyla... 40 7e-04
TC9068 similar to UP|SCWA_YEAST (Q04951) Probable family 17 gluc... 39 8e-04
TC12959 similar to UP|AAS52042 (AAS52042) ADR122Cp, partial (6%) 39 8e-04
BF177554 39 0.001
AV416122 39 0.001
TC11270 similar to UP|Q9LM88 (Q9LM88) F2D10.15, partial (9%) 39 0.001
BP036655 39 0.001
TC18665 similar to UP|O23061 (O23061) BAC IG005I10, partial (65%) 38 0.002
>TC17157 similar to UP|CAD27462 (CAD27462) Nucleosome assembly protein
1-like protein 3, partial (81%)
Length = 1298
Score = 358 bits (919), Expect = e-100
Identities = 177/303 (58%), Positives = 227/303 (74%), Gaps = 14/303 (4%)
Frame = +2
Query: 19 SPIVEKRLEVLKEIQKEHDDLKAKFFEERAALEAKYQLLYQPLYTKRYEIVNGLAEVEGA 78
SP V KR+EVL++IQ EHD+L+AKF EERA LEAKYQ LY+PLY KRYEIVNG+ EVEG
Sbjct: 2 SPEVRKRVEVLRDIQGEHDELEAKFLEERAQLEAKYQKLYEPLYKKRYEIVNGIVEVEGV 181
Query: 79 DIDEP-------EVKGVPYFWLVALQNNDEIADEITERDEDALKYLKDIKYTRTTEPQGF 131
+ E KGVP FWL A++ N+ +A+EITERDE ALKYL+DIK+ + EP+GF
Sbjct: 182 TNEAASEEDKAAEEKGVPNFWLNAMKTNETLAEEITERDEGALKYLQDIKWCKVDEPKGF 361
Query: 132 KLEFFFDSNPYFSNTILTKTYHMVDEDEFIMEKAIGTEIEWLPGKSLTE-------KKDP 184
KL+F+FDSNPYF N++LTKTYHM+DE++ I+EKAIGTEIEW PGK LT+ KK
Sbjct: 362 KLDFYFDSNPYFKNSVLTKTYHMIDEEDPILEKAIGTEIEWHPGKCLTQKILKKKPKKGS 541
Query: 185 NTAKQTIETEKFDSFFNFFNSLEIPKDGMGVDEEADEELQNEMEHDYDIGSTIRDKIIPN 244
K +TEK +SFFNFFN ++P+D +D++A EELQN MEHDYDIGSTIRDKIIP+
Sbjct: 542 KNTKPITKTEKCESFFNFFNPPQVPEDDDDIDDDAVEELQNLMEHDYDIGSTIRDKIIPH 721
Query: 245 AVSWYTGEASDGESGDLDDDDDDEDEEDDEEEVEVEKEEEDDNYGLDDDEEDDNEDVNNP 304
AVSW+TGEA + D+++DD+D DE++DE++ + E+EE+D DDD+EDD E
Sbjct: 722 AVSWFTGEAEQSDFEDIEEDDEDGDEDEDEDDDDDEEEEDD-----DDDDEDDEEGEGKS 886
Query: 305 KKR 307
K +
Sbjct: 887 KSK 895
>TC12365 similar to UP|Q39892 (Q39892) Nucleosome assembly protein 1,
partial (47%)
Length = 621
Score = 185 bits (469), Expect = 1e-47
Identities = 92/134 (68%), Positives = 110/134 (81%), Gaps = 2/134 (1%)
Frame = +1
Query: 20 PIVEKRLEVLKEIQKEHDDLKAKFFEERAALEAKYQLLYQPLYTKRYEIVNGLAEVEGAD 79
P V KR+EVL+EIQ +HD+ +AKF EERAALEAKYQ LYQPLYTKR+EIVNG+ EVEG
Sbjct: 223 PNVRKRVEVLREIQGQHDEFEAKFLEERAALEAKYQSLYQPLYTKRFEIVNGVTEVEGG- 399
Query: 80 IDEPEVK--GVPYFWLVALQNNDEIADEITERDEDALKYLKDIKYTRTTEPQGFKLEFFF 137
DEP K GVP FWL A+ NND + +EI+E DE ALK+LKDIK++R +P+GF LEFFF
Sbjct: 400 ADEPAAKEKGVPSFWLNAMNNNDVLTEEISEHDEGALKFLKDIKWSRIDDPKGF*LEFFF 579
Query: 138 DSNPYFSNTILTKT 151
DSNPYFSN++LTKT
Sbjct: 580 DSNPYFSNSVLTKT 621
>TC17434 similar to UP|Q94K07 (Q94K07) Nucleosome assembly protein, partial
(35%)
Length = 538
Score = 127 bits (318), Expect = 3e-30
Identities = 65/113 (57%), Positives = 86/113 (75%), Gaps = 6/113 (5%)
Frame = +3
Query: 2 IQALTQQRYKALQYLFASPIVEKRLEVLKEIQKEHDDLKAKFFEERAALEAKYQLLYQPL 61
IQ+L Q L+ L SP+V KR+++L+EIQ +HD+L+A FFEERAALEAKYQ LYQPL
Sbjct: 204 IQSLAGQHSDILESL--SPVVRKRVDILREIQGQHDELEASFFEERAALEAKYQKLYQPL 377
Query: 62 YTKRYEIVNGLAEVEGA------DIDEPEVKGVPYFWLVALQNNDEIADEITE 108
Y+KRY+IV G+AEVEG D +E + KGVP FWL A++NN+ +A+EI+E
Sbjct: 378 YSKRYDIVTGVAEVEGVEKETSPDTEESKEKGVPDFWLTAMKNNEVLAEEISE 536
>BG662041
Length = 121
Score = 60.8 bits (146), Expect = 3e-10
Identities = 26/33 (78%), Positives = 30/33 (90%)
Frame = +3
Query: 224 QNEMEHDYDIGSTIRDKIIPNAVSWYTGEASDG 256
QN+ME DYDIGSTIRDKI+P+AVSW+TGEA G
Sbjct: 15 QNQMEQDYDIGSTIRDKIVPHAVSWFTGEALQG 113
>TC8606 similar to UP|AAQ82688 (AAQ82688) Epa4p, partial (7%)
Length = 1008
Score = 55.1 bits (131), Expect = 1e-08
Identities = 41/110 (37%), Positives = 56/110 (50%), Gaps = 13/110 (11%)
Frame = -2
Query: 211 DGMGVDEEADEELQNEMEHDYDIGSTIRD--KIIPN------AVSWYTGEASDGESGDLD 262
DG G E +E+L +E Y S + K P A GE + E G+
Sbjct: 578 DGDGPFGEGEEDLSSEDGAGYGNNSNNKSNSKKAPEGGPGAGAGPEENGEDEEDEDGEDQ 399
Query: 263 DDDDDEDEEDDE-----EEVEVEKEEEDDNYGLDDDEEDDNEDVNNPKKR 307
+DDDD+DE+DDE E+ E E EE+DN ++DEED+ E + PKKR
Sbjct: 398 EDDDDDDEDDDEEDDGGEDEEEEGVEEEDNEDEEEDEEDE-EALQPPKKR 252
Score = 28.9 bits (63), Expect = 1.1
Identities = 22/91 (24%), Positives = 32/91 (34%), Gaps = 48/91 (52%)
Frame = -2
Query: 255 DGESGDLDDDDDDEDEE------------------------------------------- 271
D E D +DD+ DED++
Sbjct: 626 DAEGEDGNDDEGDEDDDGDGPFGEGEEDLSSEDGAGYGNNSNNKSNSKKAPEGGPGAGAG 447
Query: 272 -----DDEEEVEVEKEEEDDNYGLDDDEEDD 297
+DEE+ + E +E+DD+ DDDEEDD
Sbjct: 446 PEENGEDEEDEDGEDQEDDDDDDEDDDEEDD 354
>BP034655
Length = 517
Score = 50.8 bits (120), Expect = 3e-07
Identities = 20/39 (51%), Positives = 33/39 (84%)
Frame = +1
Query: 262 DDDDDDEDEEDDEEEVEVEKEEEDDNYGLDDDEEDDNED 300
DD++++E+EE++EEE E E+EEE++ +DDEED++ED
Sbjct: 139 DDEEEEEEEEEEEEEEEEEEEEEEEEEDEEDDEEDEDED 255
Score = 50.1 bits (118), Expect = 5e-07
Identities = 22/43 (51%), Positives = 31/43 (71%)
Frame = +1
Query: 258 SGDLDDDDDDEDEEDDEEEVEVEKEEEDDNYGLDDDEEDDNED 300
+ D DDDE+EE++EEE E E+EEE++ ++DEEDD ED
Sbjct: 115 AADFGLSDDDEEEEEEEEEEEEEEEEEEEEEEEEEDEEDDEED 243
>TC11009 similar to GB|AAP21377.1|30102918|BT006569 At1g47970 {Arabidopsis
thaliana;}, partial (44%)
Length = 700
Score = 49.3 bits (116), Expect = 8e-07
Identities = 31/108 (28%), Positives = 49/108 (44%), Gaps = 10/108 (9%)
Frame = +2
Query: 211 DGMGVDEEADEELQNEMEHDYDIGSTIRDKIIPNAVSWYTGEASDGESGDLDDDDDDEDE 270
D G D++ DE+ ++ D D+ V G D + D DDDDDDE+E
Sbjct: 38 DDDGEDDDGDEDDDDDAPGGGDDDDDEEDEEEEGGVEGGRGGGGDPDDDDDDDDDDDEEE 217
Query: 271 EDDEE----------EVEVEKEEEDDNYGLDDDEEDDNEDVNNPKKRV 308
E++E+ E EE ++ ++DE DDN++ + K V
Sbjct: 218 EEEEDLGTEYLVRRTVAAAEDEEASSDFEPEEDEGDDNDNDDGEKAGV 361
Score = 45.4 bits (106), Expect = 1e-05
Identities = 26/93 (27%), Positives = 45/93 (47%), Gaps = 1/93 (1%)
Frame = +2
Query: 216 DEEADEELQNEMEHDYDIGSTIRDKIIPNAVSWYTGEASDGESGDLDDDDDDEDEEDDEE 275
D++ D++ E E + D+G+ + ++ V+ E + + +D+ DD D +D E+
Sbjct: 182 DDDDDDDDDEEEEEEEDLGT---EYLVRRTVAAAEDEEASSDFEPEEDEGDDNDNDDGEK 352
Query: 276 EVEVEKEEEDDNYGL-DDDEEDDNEDVNNPKKR 307
K + D G DDD +D ED P KR
Sbjct: 353 AGVPSKRKRSDKDGSGDDDSDDGGEDDERPSKR 451
Score = 42.7 bits (99), Expect = 8e-05
Identities = 21/43 (48%), Positives = 28/43 (64%)
Frame = +2
Query: 258 SGDLDDDDDDEDEEDDEEEVEVEKEEEDDNYGLDDDEEDDNED 300
SGD DDDDD ED++ DE++ + DD DDDEED+ E+
Sbjct: 20 SGDEDDDDDGEDDDGDEDDDDDAPGGGDD----DDDEEDEEEE 136
Score = 38.9 bits (89), Expect = 0.001
Identities = 23/70 (32%), Positives = 40/70 (56%), Gaps = 13/70 (18%)
Frame = +2
Query: 244 NAVSWYTGEASDGESGDLDDDDDDEDEED------DEEEVEVEKEEEDDNYG-------L 290
N+ + +G+ D + G+ DDD DED++D D+++ E ++EEE G
Sbjct: 2 NSRTRMSGDEDDDDDGE--DDDGDEDDDDDAPGGGDDDDDEEDEEEEGGVEGGRGGGGDP 175
Query: 291 DDDEEDDNED 300
DDD++DD++D
Sbjct: 176 DDDDDDDDDD 205
>TC13318 similar to UP|CAA20314 (CAA20314) SPBC30B4.01c protein (SPBC3D6.14C
protein) (Fragment), partial (15%)
Length = 543
Score = 47.0 bits (110), Expect = 4e-06
Identities = 23/50 (46%), Positives = 35/50 (70%)
Frame = +3
Query: 252 EASDGESGDLDDDDDDEDEEDDEEEVEVEKEEEDDNYGLDDDEEDDNEDV 301
E D E D D+DDDD+D++DD+ E E+E++DD DD+EE+ +E+V
Sbjct: 261 EDDDEEEDDEDEDDDDDDDDDDDGE---EEEDDDD----DDEEEEGSEEV 389
Score = 43.5 bits (101), Expect = 5e-05
Identities = 15/47 (31%), Positives = 34/47 (71%)
Frame = +3
Query: 262 DDDDDDEDEEDDEEEVEVEKEEEDDNYGLDDDEEDDNEDVNNPKKRV 308
+++DDDE+E+D++E+ + + +++DD +DD++DD E+ + + V
Sbjct: 255 EEEDDDEEEDDEDEDDDDDDDDDDDGEEEEDDDDDDEEEEGSEEVEV 395
Score = 42.0 bits (97), Expect = 1e-04
Identities = 16/46 (34%), Positives = 33/46 (70%)
Frame = +3
Query: 255 DGESGDLDDDDDDEDEEDDEEEVEVEKEEEDDNYGLDDDEEDDNED 300
DG + D+++E+++D+EE+ E E +++DD+ D +EE+D++D
Sbjct: 219 DGPPKPVQTDNEEEEDDDEEEDDEDEDDDDDDDDDDDGEEEEDDDD 356
Score = 41.2 bits (95), Expect = 2e-04
Identities = 12/47 (25%), Positives = 36/47 (76%)
Frame = +3
Query: 262 DDDDDDEDEEDDEEEVEVEKEEEDDNYGLDDDEEDDNEDVNNPKKRV 308
+++++D+DEE+D+E+ + + +++DD+ G +++++DD+++ + V
Sbjct: 249 NEEEEDDDEEEDDEDEDDDDDDDDDDDGEEEEDDDDDDEEEEGSEEV 389
Score = 37.7 bits (86), Expect = 0.002
Identities = 17/40 (42%), Positives = 25/40 (62%)
Frame = +3
Query: 252 EASDGESGDLDDDDDDEDEEDDEEEVEVEKEEEDDNYGLD 291
E D + D DDDD +E+E+DD+++ E E EE + G D
Sbjct: 288 EDEDDDDDDDDDDDGEEEEDDDDDDEEEEGSEEVEVEGKD 407
Score = 33.9 bits (76), Expect = 0.036
Identities = 22/74 (29%), Positives = 36/74 (47%)
Frame = +3
Query: 216 DEEADEELQNEMEHDYDIGSTIRDKIIPNAVSWYTGEASDGESGDLDDDDDDEDEEDDEE 275
+E+ DEE +E E D D + D + G+ +++DDD+D+E++E
Sbjct: 258 EEDDDEEEDDEDEDDDD-------------------DDDDDDDGE-EEEDDDDDDEEEEG 377
Query: 276 EVEVEKEEEDDNYG 289
EVE E +D G
Sbjct: 378 SEEVEVEGKDGAKG 419
>BP048490
Length = 452
Score = 42.4 bits (98), Expect = 1e-04
Identities = 23/92 (25%), Positives = 40/92 (43%)
Frame = +3
Query: 217 EEADEELQNEMEHDYDIGSTIRDKIIPNAVSWYTGEASDGESGDLDDDDDDEDEEDDEEE 276
E +D+E + + D P+AV D DDED D + E
Sbjct: 18 ESSDDESSESSDEENDSKPATTTVAKPSAVK-----------KSASSDSDDEDSSDSDSE 164
Query: 277 VEVEKEEEDDNYGLDDDEEDDNEDVNNPKKRV 308
V+ +K + D++ +D +E+D + P+K+V
Sbjct: 165 VKKDKMDVDESDSSEDSDEEDEKSTKTPQKKV 260
>TC9200 similar to UP|Q8LLE6 (Q8LLE6) RNA-binding protein AKIP1, partial
(31%)
Length = 727
Score = 42.0 bits (97), Expect = 1e-04
Identities = 23/79 (29%), Positives = 42/79 (53%)
Frame = +1
Query: 208 IPKDGMGVDEEADEELQNEMEHDYDIGSTIRDKIIPNAVSWYTGEASDGESGDLDDDDDD 267
+PK E +LQN++E + + +++ E + E + ++ +++
Sbjct: 124 VPKSSQSA--ELPFKLQNKLEAEVQEYEEVEEEVEEEV------EEEEEEEEEEEEGEEE 279
Query: 268 EDEEDDEEEVEVEKEEEDD 286
E+EE++EEEVE E EEEDD
Sbjct: 280 EEEEEEEEEVEAEAEEEDD 336
Score = 37.4 bits (85), Expect = 0.003
Identities = 14/35 (40%), Positives = 27/35 (77%)
Frame = +1
Query: 263 DDDDDEDEEDDEEEVEVEKEEEDDNYGLDDDEEDD 297
+++++E+EE++E E E E+EEE++ + +EEDD
Sbjct: 232 EEEEEEEEEEEEGEEEEEEEEEEEEVEAEAEEEDD 336
Score = 35.8 bits (81), Expect = 0.009
Identities = 15/41 (36%), Positives = 32/41 (77%), Gaps = 1/41 (2%)
Frame = +1
Query: 262 DDDDDDEDEEDDEEEVEVEKEEEDDNYGLD-DDEEDDNEDV 301
++ +++E+EE++EEE E E+EEE++ ++ + EE+D+E +
Sbjct: 223 EEVEEEEEEEEEEEEGEEEEEEEEEEEEVEAEAEEEDDEPI 345
Score = 35.4 bits (80), Expect = 0.012
Identities = 13/46 (28%), Positives = 32/46 (69%)
Frame = +1
Query: 252 EASDGESGDLDDDDDDEDEEDDEEEVEVEKEEEDDNYGLDDDEEDD 297
E + ++++++++E+EE++ EE E E+EEE++ ++E+D+
Sbjct: 202 EVEEEVEEEVEEEEEEEEEEEEGEEEEEEEEEEEEVEAEAEEEDDE 339
Score = 35.0 bits (79), Expect = 0.016
Identities = 15/34 (44%), Positives = 26/34 (76%)
Frame = +1
Query: 267 DEDEEDDEEEVEVEKEEEDDNYGLDDDEEDDNED 300
+E EE+ EEEVE E+EEE++ +++EE++ E+
Sbjct: 199 EEVEEEVEEEVEEEEEEEEEEEEGEEEEEEEEEE 300
Score = 33.5 bits (75), Expect = 0.047
Identities = 14/38 (36%), Positives = 27/38 (70%)
Frame = +1
Query: 263 DDDDDEDEEDDEEEVEVEKEEEDDNYGLDDDEEDDNED 300
+++ +E+EE++EEE E E+EEE++ + + E + ED
Sbjct: 220 EEEVEEEEEEEEEEEEGEEEEEEEEEEEEVEAEAEEED 333
Score = 33.5 bits (75), Expect = 0.047
Identities = 15/47 (31%), Positives = 32/47 (67%)
Frame = +1
Query: 255 DGESGDLDDDDDDEDEEDDEEEVEVEKEEEDDNYGLDDDEEDDNEDV 301
+ E + ++ +++ +EE +EEE E E+EEE + +++EE++ E+V
Sbjct: 178 EAEVQEYEEVEEEVEEEVEEEEEEEEEEEEGEE---EEEEEEEEEEV 309
Score = 32.3 bits (72), Expect = 0.10
Identities = 23/92 (25%), Positives = 44/92 (47%)
Frame = +1
Query: 209 PKDGMGVDEEADEELQNEMEHDYDIGSTIRDKIIPNAVSWYTGEASDGESGDLDDDDDDE 268
P + E +E + NE T + K++P S + E L+ + +
Sbjct: 46 PHSLASLQESPNEAMVNE-------SITKKRKLVPK--SSQSAELPFKLQNKLEAEVQEY 198
Query: 269 DEEDDEEEVEVEKEEEDDNYGLDDDEEDDNED 300
+E ++E E EVE+EEE++ + +EE++ E+
Sbjct: 199 EEVEEEVEEEVEEEEEEEEEEEEGEEEEEEEE 294
>BP043430
Length = 493
Score = 40.0 bits (92), Expect = 5e-04
Identities = 18/46 (39%), Positives = 25/46 (54%)
Frame = -1
Query: 255 DGESGDLDDDDDDEDEEDDEEEVEVEKEEEDDNYGLDDDEEDDNED 300
D E D+DD+ D E DEE+ E E +DD+ ++ DD ED
Sbjct: 376 DNEGSGKDNDDEGNDNEVDEEDEEEEDSNDDDDGSESEEFNDDEED 239
Score = 39.3 bits (90), Expect = 8e-04
Identities = 18/51 (35%), Positives = 33/51 (64%), Gaps = 2/51 (3%)
Frame = -1
Query: 250 TGEASDGESGDLDDD--DDDEDEEDDEEEVEVEKEEEDDNYGLDDDEEDDN 298
+G+ ++G D DD+ D++ DEED+EEE + ++ ++ +DDEED +
Sbjct: 385 SGDDNEGSGKDNDDEGNDNEVDEEDEEEEDSNDDDDGSESEEFNDDEEDSD 233
Score = 33.1 bits (74), Expect = 0.061
Identities = 16/53 (30%), Positives = 33/53 (62%)
Frame = -1
Query: 254 SDGESGDLDDDDDDEDEEDDEEEVEVEKEEEDDNYGLDDDEEDDNEDVNNPKK 306
+ G+ + D+D++ D+E + E ++EEED N DDD+ ++E+ N+ ++
Sbjct: 388 ASGDDNEGSGKDNDDEGNDNEVD-EEDEEEEDSN---DDDDGSESEEFNDDEE 242
>AI967639
Length = 335
Score = 40.0 bits (92), Expect = 5e-04
Identities = 20/48 (41%), Positives = 32/48 (66%), Gaps = 3/48 (6%)
Frame = +1
Query: 263 DDDDDEDEEDDEEEVEVEKEEEDDNYGLDDDEEDD---NEDVNNPKKR 307
+ D++++EED +E +E EEEDD DDD++DD +ED ++ K R
Sbjct: 97 EGDEEDEEEDQGDEYNLEDEEEDD----DDDDDDDLSISEDSDSDKPR 228
Score = 29.6 bits (65), Expect = 0.67
Identities = 9/35 (25%), Positives = 25/35 (70%)
Frame = +1
Query: 252 EASDGESGDLDDDDDDEDEEDDEEEVEVEKEEEDD 286
E G+ +L+D+++D+D +DD++++ + ++ + D
Sbjct: 118 EEDQGDEYNLEDEEEDDD-DDDDDDLSISEDSDSD 219
Score = 26.9 bits (58), Expect = 4.4
Identities = 10/42 (23%), Positives = 27/42 (63%), Gaps = 7/42 (16%)
Frame = +1
Query: 253 ASDGESGDLDDD-------DDDEDEEDDEEEVEVEKEEEDDN 287
+S+G+ D ++D +D+E+++DD+++ ++ E+ D+
Sbjct: 91 SSEGDEEDEEEDQGDEYNLEDEEEDDDDDDDDDLSISEDSDS 216
Score = 26.6 bits (57), Expect = 5.7
Identities = 11/28 (39%), Positives = 17/28 (60%)
Frame = +1
Query: 273 DEEEVEVEKEEEDDNYGLDDDEEDDNED 300
DEE ++ K+ D + + DEED+ ED
Sbjct: 43 DEEYLKKRKQRRDISSSSEGDEEDEEED 126
Score = 26.2 bits (56), Expect = 7.4
Identities = 11/31 (35%), Positives = 19/31 (60%)
Frame = +1
Query: 251 GEASDGESGDLDDDDDDEDEEDDEEEVEVEK 281
G+ + E + DDDDDD+D+ E+ + +K
Sbjct: 130 GDEYNLEDEEEDDDDDDDDDLSISEDSDSDK 222
>TC8507 similar to UP|Q8LJS2 (Q8LJS2) Nucleolar histone deacetylase
HD2-P39, partial (40%)
Length = 620
Score = 39.7 bits (91), Expect = 7e-04
Identities = 17/48 (35%), Positives = 28/48 (57%)
Frame = +3
Query: 260 DLDDDDDDEDEEDDEEEVEVEKEEEDDNYGLDDDEEDDNEDVNNPKKR 307
D +DD DDED+EDD+ + E++ + D D+ + D+ D P K+
Sbjct: 3 DEEDDSDDEDDEDDDSDEEMDDADSD-----SDESDSDDTDEETPVKK 131
Score = 38.1 bits (87), Expect = 0.002
Identities = 19/53 (35%), Positives = 30/53 (55%)
Frame = +3
Query: 255 DGESGDLDDDDDDEDEEDDEEEVEVEKEEEDDNYGLDDDEEDDNEDVNNPKKR 307
+ +S D DD+DDD DEE D+ + + ++ + DD DEE + + KKR
Sbjct: 9 EDDSDDEDDEDDDSDEEMDDADSDSDESDSDDT-----DEETPVKKADQGKKR 152
>TC9068 similar to UP|SCWA_YEAST (Q04951) Probable family 17 glucosidase
SCW10 precursor (Soluble cell wall protein 10) ,
partial (6%)
Length = 1209
Score = 39.3 bits (90), Expect = 8e-04
Identities = 14/35 (40%), Positives = 26/35 (74%)
Frame = +2
Query: 255 DGESGDLDDDDDDEDEEDDEEEVEVEKEEEDDNYG 289
D + D DDDDDD+D++DD+++ + +K++ D + G
Sbjct: 83 DDDDDDDDDDDDDDDDDDDDDDDDGDKKQPDSDTG 187
Score = 37.4 bits (85), Expect = 0.003
Identities = 18/43 (41%), Positives = 26/43 (59%)
Frame = +2
Query: 262 DDDDDDEDEEDDEEEVEVEKEEEDDNYGLDDDEEDDNEDVNNP 304
DDDDDD+D++DD ++DD DDD++DD+ D P
Sbjct: 83 DDDDDDDDDDDD---------DDDD----DDDDDDDDGDKKQP 172
Score = 36.6 bits (83), Expect = 0.006
Identities = 13/38 (34%), Positives = 26/38 (68%)
Frame = +2
Query: 249 YTGEASDGESGDLDDDDDDEDEEDDEEEVEVEKEEEDD 286
++ + D + D DDDDDD+D++DD+++ +K+ + D
Sbjct: 68 FSPKPDDDDDDDDDDDDDDDDDDDDDDDDGDKKQPDSD 181
Score = 35.8 bits (81), Expect = 0.009
Identities = 13/36 (36%), Positives = 24/36 (66%)
Frame = +2
Query: 255 DGESGDLDDDDDDEDEEDDEEEVEVEKEEEDDNYGL 290
D + D DDDDDD+D++DD+++ + ++ + D L
Sbjct: 89 DDDDDDDDDDDDDDDDDDDDDDGDKKQPDSDTGTAL 196
Score = 26.2 bits (56), Expect = 7.4
Identities = 9/20 (45%), Positives = 16/20 (80%)
Frame = +2
Query: 281 KEEEDDNYGLDDDEEDDNED 300
K ++DD+ DDD++DD++D
Sbjct: 77 KPDDDDDDDDDDDDDDDDDD 136
>TC12959 similar to UP|AAS52042 (AAS52042) ADR122Cp, partial (6%)
Length = 517
Score = 39.3 bits (90), Expect = 8e-04
Identities = 17/35 (48%), Positives = 24/35 (68%)
Frame = +1
Query: 254 SDGESGDLDDDDDDEDEEDDEEEVEVEKEEEDDNY 288
SDG S D DDDDDD ++EDD+ + E++DD +
Sbjct: 112 SDGSSSDDDDDDDDSEDEDDD-----DNEDDDDEW 201
>BF177554
Length = 246
Score = 38.9 bits (89), Expect = 0.001
Identities = 23/53 (43%), Positives = 33/53 (61%), Gaps = 2/53 (3%)
Frame = +1
Query: 250 TGEASDGESGDLDDDDDDEDEEDDEEEVEVEKEEEDDNYGLDD--DEEDDNED 300
T E D +S + + + E+EE++EEE E E+EEEDD G D EE++ ED
Sbjct: 10 TSEQQDEDSCEEEAGESSEEEEEEEEEGE-EEEEEDDGPGDRDAQPEEEEEED 165
Score = 33.9 bits (76), Expect = 0.036
Identities = 17/41 (41%), Positives = 26/41 (62%)
Frame = +1
Query: 257 ESGDLDDDDDDEDEEDDEEEVEVEKEEEDDNYGLDDDEEDD 297
E G + D+D EE+ E E E+EEE++ G +++EEDD
Sbjct: 1 EFGTSEQQDEDSCEEEAGESSEEEEEEEEE--GEEEEEEDD 117
>AV416122
Length = 414
Score = 38.9 bits (89), Expect = 0.001
Identities = 21/54 (38%), Positives = 30/54 (54%)
Frame = +3
Query: 253 ASDGESGDLDDDDDDEDEEDDEEEVEVEKEEEDDNYGLDDDEEDDNEDVNNPKK 306
A+D D D + E+EEDDEE+ E E EEEDD +++D N + +K
Sbjct: 258 ANDDNEPDNDVQFEGEEEEDDEEDKEEEDEEEDD------EDDDSNGEAEVDRK 401
Score = 37.7 bits (86), Expect = 0.002
Identities = 17/48 (35%), Positives = 32/48 (66%)
Frame = +3
Query: 253 ASDGESGDLDDDDDDEDEEDDEEEVEVEKEEEDDNYGLDDDEEDDNED 300
A+D S + +D++E D++ + E E+EE+D+ ++DEE+D+ED
Sbjct: 222 ATDDCSSQPKEAANDDNEPDNDVQFEGEEEEDDEEDKEEEDEEEDDED 365
Score = 32.7 bits (73), Expect = 0.080
Identities = 11/36 (30%), Positives = 23/36 (63%)
Frame = +3
Query: 249 YTGEASDGESGDLDDDDDDEDEEDDEEEVEVEKEEE 284
+ GE + + D +++D++ED+EDD+ E E + +
Sbjct: 294 FEGEEEEDDEEDKEEEDEEEDDEDDDSNGEAEVDRK 401
Score = 30.4 bits (67), Expect = 0.39
Identities = 13/33 (39%), Positives = 21/33 (63%)
Frame = +3
Query: 257 ESGDLDDDDDDEDEEDDEEEVEVEKEEEDDNYG 289
E D ++D ++EDEE+D+E+ + E E D G
Sbjct: 306 EEEDDEEDKEEEDEEEDDEDDDSNGEAEVDRKG 404
Score = 28.9 bits (63), Expect = 1.1
Identities = 19/75 (25%), Positives = 32/75 (42%), Gaps = 1/75 (1%)
Frame = +3
Query: 209 PKDGMGVDEEADEELQNEMEHDYDIGSTIRDKIIPNAVSWYTGEASDGESGDLDDDDDDE 268
PK+ D E D ++Q E E E D E ++D++++
Sbjct: 246 PKEAANDDNEPDNDVQFEGE-----------------------EEEDDEEDKEEEDEEED 356
Query: 269 DEEDDEE-EVEVEKE 282
DE+DD E EV+++
Sbjct: 357 DEDDDSNGEAEVDRK 401
Score = 27.7 bits (60), Expect = 2.6
Identities = 10/39 (25%), Positives = 23/39 (58%)
Frame = +3
Query: 252 EASDGESGDLDDDDDDEDEEDDEEEVEVEKEEEDDNYGL 290
E + E + D +++DE+E+D++++ E E + G+
Sbjct: 297 EGEEEEDDEEDKEEEDEEEDDEDDDSNGEAEVDRKGKGI 413
>TC11270 similar to UP|Q9LM88 (Q9LM88) F2D10.15, partial (9%)
Length = 735
Score = 38.5 bits (88), Expect = 0.001
Identities = 18/46 (39%), Positives = 30/46 (65%)
Frame = +3
Query: 262 DDDDDDEDEEDDEEEVEVEKEEEDDNYGLDDDEEDDNEDVNNPKKR 307
++D+++E+ E++EEE EV E+E N D+DEE++ V K R
Sbjct: 423 EEDEEEENGEEEEEEEEVVVEDEQGNEDEDEDEEEEGVVVVEVKGR 560
Score = 34.3 bits (77), Expect = 0.027
Identities = 17/63 (26%), Positives = 36/63 (56%), Gaps = 16/63 (25%)
Frame = +3
Query: 262 DDDDDDEDE----------------EDDEEEVEVEKEEEDDNYGLDDDEEDDNEDVNNPK 305
++D++DE+E E+DEEE E+EEE++ ++D++ +++ED + +
Sbjct: 348 EEDEEDEEEQTPHPTRGRSRVEHAQEEDEEEENGEEEEEEEEVVVEDEQGNEDEDEDEEE 527
Query: 306 KRV 308
+ V
Sbjct: 528 EGV 536
Score = 33.5 bits (75), Expect = 0.047
Identities = 13/23 (56%), Positives = 20/23 (86%)
Frame = +2
Query: 261 LDDDDDDEDEEDDEEEVEVEKEE 283
LD++D+DEDE++D++E EKEE
Sbjct: 236 LDEEDEDEDEDEDDDERGREKEE 304
Score = 30.4 bits (67), Expect = 0.39
Identities = 12/24 (50%), Positives = 18/24 (75%)
Frame = +2
Query: 260 DLDDDDDDEDEEDDEEEVEVEKEE 283
D +D+D+DEDE+DDE E E+ +
Sbjct: 239 DEEDEDEDEDEDDDERGREKEEAQ 310
Score = 29.6 bits (65), Expect = 0.67
Identities = 11/26 (42%), Positives = 20/26 (76%)
Frame = +2
Query: 264 DDDDEDEEDDEEEVEVEKEEEDDNYG 289
D++DEDE++DE++ E +E+E+ G
Sbjct: 239 DEEDEDEDEDEDDDERGREKEEAQAG 316
Score = 28.5 bits (62), Expect = 1.5
Identities = 18/55 (32%), Positives = 27/55 (48%)
Frame = +2
Query: 235 STIRDKIIPNAVSWYTGEASDGESGDLDDDDDDEDEEDDEEEVEVEKEEEDDNYG 289
ST+R P V Y + D E D D+D+DD++ ++EE + E E G
Sbjct: 182 STLRRSRRPRNVR-YNIDYLDEEDEDEDEDEDDDERGREKEEAQAG*EAEPGGGG 343
>BP036655
Length = 556
Score = 38.5 bits (88), Expect = 0.001
Identities = 19/41 (46%), Positives = 26/41 (63%)
Frame = -3
Query: 257 ESGDLDDDDDDEDEEDDEEEVEVEKEEEDDNYGLDDDEEDD 297
E L DD DD++ D+EEEV E ++DD ++DDE DD
Sbjct: 410 EEEGLSDDGDDDEVVDEEEEVVEEWSDDDD---INDDESDD 297
Score = 38.5 bits (88), Expect = 0.001
Identities = 17/36 (47%), Positives = 27/36 (74%), Gaps = 1/36 (2%)
Frame = -3
Query: 262 DDDDDDEDEEDDEEEVEVEK-EEEDDNYGLDDDEED 296
DD+ DDE EE+ E E+E EK EEE++ G+++ E++
Sbjct: 314 DDESDDEVEEESETEIETEKEEEEEEECGVENVEDE 207
Score = 35.0 bits (79), Expect = 0.016
Identities = 18/52 (34%), Positives = 29/52 (55%)
Frame = -3
Query: 252 EASDGESGDLDDDDDDEDEEDDEEEVEVEKEEEDDNYGLDDDEEDDNEDVNN 303
E D E +++ DD+D DDE + EVE+E E + ++EE++ V N
Sbjct: 374 EVVDEEEEVVEEWSDDDDINDDESDDEVEEESETEIETEKEEEEEEECGVEN 219
Score = 27.3 bits (59), Expect = 3.3
Identities = 14/37 (37%), Positives = 21/37 (55%), Gaps = 2/37 (5%)
Frame = -3
Query: 266 DDEDEEDDEEEVEVEKEEED--DNYGLDDDEEDDNED 300
++E DD ++ EV EEE+ + + DDD DD D
Sbjct: 410 EEEGLSDDGDDDEVVDEEEEVVEEWSDDDDINDDESD 300
>TC18665 similar to UP|O23061 (O23061) BAC IG005I10, partial (65%)
Length = 451
Score = 38.1 bits (87), Expect = 0.002
Identities = 21/58 (36%), Positives = 29/58 (49%)
Frame = -2
Query: 250 TGEASDGESGDLDDDDDDEDEEDDEEEVEVEKEEEDDNYGLDDDEEDDNEDVNNPKKR 307
+GE + E D D D+E +DDE E E E E ++ G D DE + E +KR
Sbjct: 210 SGEGDEDEDLDSDGFRDEEIGDDDEGEGEEEDEATEEASGFDGDEHEVRERAPGDEKR 37
Database: LJGI
Posted date: Jul 30, 2004 11:16 AM
Number of letters in database: 14,692,800
Number of sequences in database: 28,460
Lambda K H
0.310 0.134 0.374
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 3,990,475
Number of Sequences: 28460
Number of extensions: 48451
Number of successful extensions: 2710
Number of sequences better than 10.0: 253
Number of HSP's better than 10.0 without gapping: 786
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1593
length of query: 308
length of database: 4,897,600
effective HSP length: 90
effective length of query: 218
effective length of database: 2,336,200
effective search space: 509291600
effective search space used: 509291600
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.7 bits)
S2: 55 (25.8 bits)
Medicago: description of AC138526.4


