

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC138451.8 - phase: 0
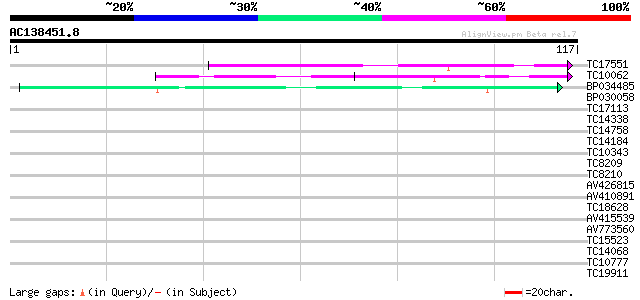
(117 letters)
Database: LJGI
28,460 sequences; 14,692,800 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC17551 weakly similar to UP|AAR24662 (AAR24662) At5g26960, part... 40 1e-04
TC10062 similar to BAD08700 (BAD08700) Cold shock domain protein... 40 1e-04
BP034485 37 9e-04
BP030058 37 0.001
TC17113 weakly similar to UP|Q9LFU8 (Q9LFU8) Proline-rich protei... 36 0.001
TC14338 similar to UP|Q9MAH5 (Q9MAH5) F12M16.16, partial (33%) 36 0.001
TC14758 similar to UP|O24601 (O24601) Glycine-rich RNA binding p... 36 0.002
TC14184 similar to UP|Q93WZ6 (Q93WZ6) Abscisic stress ripening-l... 35 0.003
TC10343 similar to UP|Q9SZZ1 (Q9SZZ1) Fibrillarin-like protein (... 35 0.003
TC8209 homologue to PIR|T10586|T10586 small nuclear ribonucleopr... 34 0.006
TC8210 homologue to PIR|T10586|T10586 small nuclear ribonucleopr... 34 0.006
AV426815 33 0.010
AV410891 33 0.012
TC18628 weakly similar to PIR|T09527|T09527 glycine-rich protein... 33 0.012
AV415539 33 0.012
AV773560 33 0.016
TC15523 similar to UP|GRPA_MAIZE (P10979) Glycine-rich RNA-bindi... 33 0.016
TC14068 homologue to UP|Q9SLR7 (Q9SLR7) Thiamin biosynthetic enz... 32 0.021
TC10777 similar to UP|Q9SCL3 (Q9SCL3) Pre-mRNA splicing factor S... 32 0.028
TC19911 weakly similar to PIR|A44805|A44805 eggshell protein pre... 32 0.028
>TC17551 weakly similar to UP|AAR24662 (AAR24662) At5g26960, partial (22%)
Length = 505
Score = 40.0 bits (92), Expect = 1e-04
Identities = 29/80 (36%), Positives = 34/80 (42%), Gaps = 5/80 (6%)
Frame = -3
Query: 42 KGNEGKSNDGCGDGIEIHCRKGYVDGTIWDKDISITKGTGWQTTGRKG-----GGSGKGG 96
+G G+ G G R G G+IW + GW+ GR G SG GG
Sbjct: 428 RGARGRGGSGRSRGRRRCSRGGRGRGSIWRRG-------GWRRRGRSRWR*GRGRSGGGG 270
Query: 97 KGGGGGYRIPIPGVGKGGGG 116
GGGGG R G GGGG
Sbjct: 269 GGGGGGRR----SRGSGGGG 222
Score = 26.2 bits (56), Expect = 1.5
Identities = 24/88 (27%), Positives = 32/88 (36%), Gaps = 6/88 (6%)
Frame = -1
Query: 35 ECGGAILKGNEGKSNDGCGDGIEIHCRKGYVDGTIWDKDISITKGTGWQTTGRKGGGS-- 92
E GG + G+ G + + G V G + +G G + G G
Sbjct: 334 EGGGGEAEVGGGEGEGGVAEEVAAAAEVGEVGGVEEVGPTAADEGEGREGRGWDAGEEVE 155
Query: 93 ----GKGGKGGGGGYRIPIPGVGKGGGG 116
+GG GGG R G G GGGG
Sbjct: 154 NDVVREGGNGGGRLLRWFGDGGGGGGGG 71
>TC10062 similar to BAD08700 (BAD08700) Cold shock domain protein 2, partial
(66%)
Length = 481
Score = 39.7 bits (91), Expect = 1e-04
Identities = 32/86 (37%), Positives = 35/86 (40%)
Frame = +3
Query: 31 RNTNECGGAILKGNEGKSNDGCGDGIEIHCRKGYVDGTIWDKDISITKGTGWQTTGRKGG 90
+ T GGA G G+ G G G GY G G G G GG
Sbjct: 264 QGTRRGGGA---GGGGRGGGGYGGG-------GYGGGGYGGG------GRGGGGYGGGGG 395
Query: 91 GSGKGGKGGGGGYRIPIPGVGKGGGG 116
G GG GGGGGY G G+GGGG
Sbjct: 396 YGGSGGYGGGGGYGGGGRGGGRGGGG 473
Score = 37.0 bits (84), Expect = 9e-04
Identities = 23/46 (50%), Positives = 27/46 (58%), Gaps = 1/46 (2%)
Frame = +3
Query: 72 KDISITKGTGWQTTG-RKGGGSGKGGKGGGGGYRIPIPGVGKGGGG 116
K I +T G G R+GGG+G GG+ GGGGY G G GGGG
Sbjct: 225 KAIEVTGPNGQAVQGTRRGGGAGGGGR-GGGGY----GGGGYGGGG 347
Score = 28.5 bits (62), Expect = 0.31
Identities = 12/22 (54%), Positives = 14/22 (63%)
Frame = +3
Query: 81 GWQTTGRKGGGSGKGGKGGGGG 102
G+ G GGG GG+GGGGG
Sbjct: 411 GYGGGGGYGGGGRGGGRGGGGG 476
Score = 25.8 bits (55), Expect = 2.0
Identities = 11/17 (64%), Positives = 11/17 (64%)
Frame = +3
Query: 86 GRKGGGSGKGGKGGGGG 102
G GGG G G GGGGG
Sbjct: 429 GYGGGGRGGGRGGGGGG 479
>BP034485
Length = 485
Score = 37.0 bits (84), Expect = 9e-04
Identities = 39/138 (28%), Positives = 54/138 (38%), Gaps = 26/138 (18%)
Frame = +3
Query: 3 KQKKSKINIGVHEYANWLGKGTLLQEG-------------DRNTNECGGAILKGNEGKSN 49
K+KK+K G +LG G ++G ++N N GG KG EGK
Sbjct: 69 KKKKTKDKNGGGFLVKFLGLGKKSKKGGGGGGGETANKNKNKNKNN-GGGNNKGKEGKKG 245
Query: 50 DGCGDGIEIHCRKGYVDGTIWDKDISITKGTGWQTTGRKGGGSGKGGK------------ 97
G G ++ +D D DI +G G+ GGG GKG K
Sbjct: 246 GGGGGKLD------KIDFDFQDFDIPSHGKSG----GKGGGGGGKGNKNGHDHGHGIGGN 395
Query: 98 -GGGGGYRIPIPGVGKGG 114
G GG+ P+ +G G
Sbjct: 396 MNGNGGHMDPMSQLGSMG 449
>BP030058
Length = 455
Score = 36.6 bits (83), Expect = 0.001
Identities = 20/46 (43%), Positives = 23/46 (49%)
Frame = +1
Query: 71 DKDISITKGTGWQTTGRKGGGSGKGGKGGGGGYRIPIPGVGKGGGG 116
DK + G+ G +GGG G G GGGGG G G GGGG
Sbjct: 259 DKGLQFCGQEGYFGGGGEGGGGGGGDGGGGGG-----DGHGSGGGG 381
Score = 27.3 bits (59), Expect = 0.69
Identities = 24/76 (31%), Positives = 26/76 (33%), Gaps = 2/76 (2%)
Frame = +1
Query: 29 GDRNTNECG--GAILKGNEGKSNDGCGDGIEIHCRKGYVDGTIWDKDISITKGTGWQTTG 86
GD+ CG G G EG G GDG G G
Sbjct: 256 GDKGLQFCGQEGYFGGGGEGGGGGG-GDG-----------------------GGGGGDGH 363
Query: 87 RKGGGSGKGGKGGGGG 102
GGG G+G GGGGG
Sbjct: 364 GSGGGGGEGDGGGGGG 411
>TC17113 weakly similar to UP|Q9LFU8 (Q9LFU8) Proline-rich protein, partial
(25%)
Length = 1077
Score = 36.2 bits (82), Expect = 0.001
Identities = 34/97 (35%), Positives = 41/97 (42%), Gaps = 6/97 (6%)
Frame = -2
Query: 26 LQEGDRNTNECGGAILKGN---EGKSNDGCGDGIEIHCRKGYVDGTIWDK-DISITKGTG 81
L +G+ E GG GN +G N G G G + G G +K + G G
Sbjct: 854 LDQGEEFFVEIGGLPGGGNNGGKGGKNGGGGGGGGGGVKPGITIGGKGNKGEGGCFDGGG 675
Query: 82 WQTTGRKGGGSGKG--GKGGGGGYRIPIPGVGKGGGG 116
W G GGG G G G G GG G+GK GGG
Sbjct: 674 WNGLGINGGGLGGG*NGLGISGGGLGG*NGLGKSGGG 564
Score = 28.9 bits (63), Expect = 0.24
Identities = 17/37 (45%), Positives = 20/37 (53%), Gaps = 1/37 (2%)
Frame = -2
Query: 81 GWQTTGRKGGGSGKGGKG-GGGGYRIPIPGVGKGGGG 116
GW +G+ GGG G G G GGG G+G GGG
Sbjct: 557 GWNGSGKSGGGLG*KGLGIRGGG*N----GLGINGGG 459
Score = 25.0 bits (53), Expect = 3.4
Identities = 28/82 (34%), Positives = 36/82 (43%), Gaps = 7/82 (8%)
Frame = -2
Query: 42 KGNEGKSNDGCGDG---IEIHCRKGYVDGTIWDKDISITKGTGWQTTGRKGGG----SGK 94
KGN+G+ GC DG + G + G IS G G+ GGG +G
Sbjct: 716 KGNKGEG--GCFDGGGWNGLGINGGGLGGG*NGLGISGGGLGG*NGLGKSGGGLGGWNGS 543
Query: 95 GGKGGGGGYRIPIPGVGKGGGG 116
G GGG G + G+G GGG
Sbjct: 542 GKSGGGLG*K----GLGIRGGG 489
>TC14338 similar to UP|Q9MAH5 (Q9MAH5) F12M16.16, partial (33%)
Length = 1751
Score = 36.2 bits (82), Expect = 0.001
Identities = 19/56 (33%), Positives = 27/56 (47%)
Frame = -2
Query: 61 RKGYVDGTIWDKDISITKGTGWQTTGRKGGGSGKGGKGGGGGYRIPIPGVGKGGGG 116
+ G++ GT+ + I +G GG G GG+G GGG + G G GG G
Sbjct: 301 QNGFLRGTVGPVRVRIVEG----------GGGGAGGEGAGGGEEVAAGGDGYGGEG 164
Score = 27.3 bits (59), Expect = 0.69
Identities = 18/46 (39%), Positives = 21/46 (45%), Gaps = 9/46 (19%)
Frame = -1
Query: 79 GTGWQTTG---------RKGGGSGKGGKGGGGGYRIPIPGVGKGGG 115
G GW + G R GSG+ G+GGG I G G GGG
Sbjct: 284 GDGWTS*G*DR*GRRWWRGWRGSGRRGRGGGRWGWIRRRGDGSGGG 147
>TC14758 similar to UP|O24601 (O24601) Glycine-rich RNA binding protein 2,
partial (93%)
Length = 725
Score = 35.8 bits (81), Expect = 0.002
Identities = 21/40 (52%), Positives = 23/40 (57%), Gaps = 2/40 (5%)
Frame = +1
Query: 79 GTGWQTTGRKGGGSGK--GGKGGGGGYRIPIPGVGKGGGG 116
G G + G GGG G+ GG GGGGG R G G GGGG
Sbjct: 334 GGGGRGGGGYGGGGGRREGGYGGGGGSR----GYGGGGGG 441
Score = 34.7 bits (78), Expect = 0.004
Identities = 29/85 (34%), Positives = 34/85 (39%)
Frame = +1
Query: 32 NTNECGGAILKGNEGKSNDGCGDGIEIHCRKGYVDGTIWDKDISITKGTGWQTTGRKGGG 91
N NE G + NE ++ G G G G G G G + G GGG
Sbjct: 268 NGNELDGRNITVNEAQARGGGGGG------GGRGGG-------GYGGGGGRREGGYGGGG 408
Query: 92 SGKGGKGGGGGYRIPIPGVGKGGGG 116
+G GGGGGY G GGGG
Sbjct: 409 GSRGYGGGGGGY-------GGGGGG 462
Score = 34.3 bits (77), Expect = 0.006
Identities = 18/32 (56%), Positives = 19/32 (59%)
Frame = +1
Query: 86 GRKGGGSGKGGKGGGGGYRIPIPGVGKGGGGK 117
G GGG G GG GGGGG R G G GGG +
Sbjct: 328 GGGGGGRGGGGYGGGGGRR--EGGYGGGGGSR 417
Score = 32.3 bits (72), Expect = 0.021
Identities = 18/38 (47%), Positives = 19/38 (49%)
Frame = +1
Query: 79 GTGWQTTGRKGGGSGKGGKGGGGGYRIPIPGVGKGGGG 116
G G + G GGG G GG GGGG G G GGG
Sbjct: 397 GGGGGSRGYGGGGGGYGGGGGGGYGGRREGGYGGDGGG 510
Score = 29.6 bits (65), Expect = 0.14
Identities = 30/82 (36%), Positives = 34/82 (40%), Gaps = 2/82 (2%)
Frame = +1
Query: 37 GGAILKGNEGKSNDGCGDGIEIHCRKGYVDGTIWDKDISITKGTGWQTTGRKG-GGSGKG 95
GG G G+ G G G +GY G G G+ G G GG +G
Sbjct: 349 GGGGYGGGGGRREGGYGGG---GGSRGYGGG-----------GGGYGGGGGGGYGGRREG 486
Query: 96 GKGG-GGGYRIPIPGVGKGGGG 116
G GG GGG R G G GGGG
Sbjct: 487 GYGGDGGGSRY---GSGGGGGG 543
>TC14184 similar to UP|Q93WZ6 (Q93WZ6) Abscisic stress ripening-like
protein, partial (57%)
Length = 1141
Score = 35.4 bits (80), Expect = 0.003
Identities = 20/42 (47%), Positives = 22/42 (51%), Gaps = 4/42 (9%)
Frame = +3
Query: 80 TGWQTTGRKGGGSGKGGKGG----GGGYRIPIPGVGKGGGGK 117
T + T GGG G GG GG GGGY G G GGGG+
Sbjct: 339 TAYSTDEPSGGGYGTGGAGGGYSTGGGY-----GTGGGGGGE 449
Score = 32.0 bits (71), Expect = 0.028
Identities = 19/42 (45%), Positives = 21/42 (49%), Gaps = 4/42 (9%)
Frame = +3
Query: 79 GTGWQTTGRKGGGSGKGGK----GGGGGYRIPIPGVGKGGGG 116
G G+ T G G G G GG+ GGGGGY GGGG
Sbjct: 393 GGGYSTGGGYGTGGGGGGEYSTGGGGGGY-------STGGGG 497
>TC10343 similar to UP|Q9SZZ1 (Q9SZZ1) Fibrillarin-like protein (Fibrillarin
2) (AtFib2), partial (85%)
Length = 1116
Score = 35.4 bits (80), Expect = 0.003
Identities = 18/39 (46%), Positives = 23/39 (58%), Gaps = 1/39 (2%)
Frame = +1
Query: 79 GTGWQTTGRKGGGSGKG-GKGGGGGYRIPIPGVGKGGGG 116
G G++ G +G G G G G+GGG G G G+GGGG
Sbjct: 61 GGGFRGRGDRGRGRGDGGGRGGGRGTPFKARGGGRGGGG 177
Score = 34.3 bits (77), Expect = 0.006
Identities = 16/32 (50%), Positives = 20/32 (62%)
Frame = +1
Query: 86 GRKGGGSGKGGKGGGGGYRIPIPGVGKGGGGK 117
G +GGG G+GG+GGG G G+GGG K
Sbjct: 157 GGRGGGGGRGGRGGGRG--------GRGGGMK 228
Score = 30.4 bits (67), Expect = 0.081
Identities = 19/45 (42%), Positives = 22/45 (48%), Gaps = 6/45 (13%)
Frame = +1
Query: 78 KGTGWQTTGRKGGGSGK------GGKGGGGGYRIPIPGVGKGGGG 116
+G G G +GGG G GG+GGGGG G G GGG
Sbjct: 88 RGRGRGDGGGRGGGRGTPFKARGGGRGGGGGRGGRGGGRGGRGGG 222
Score = 24.6 bits (52), Expect = 4.4
Identities = 12/22 (54%), Positives = 15/22 (67%), Gaps = 1/22 (4%)
Frame = +1
Query: 96 GKGGGGGYR-IPIPGVGKGGGG 116
G+GGGGG+R G G+G GG
Sbjct: 49 GRGGGGGFRGRGDRGRGRGDGG 114
>TC8209 homologue to PIR|T10586|T10586 small nuclear
ribonucleoprotein-associated protein homolog
F9F13.90 - Arabidopsis thaliana {Arabidopsis
thaliana;}, partial (19%)
Length = 635
Score = 34.3 bits (77), Expect = 0.006
Identities = 18/30 (60%), Positives = 19/30 (63%)
Frame = -2
Query: 86 GRKGGGSGKGGKGGGGGYRIPIPGVGKGGG 115
GR GGG G G+GGGGG I PG GGG
Sbjct: 145 GRGGGGPGIPGRGGGGGPLIICPG---GGG 65
Score = 33.5 bits (75), Expect = 0.010
Identities = 19/39 (48%), Positives = 22/39 (55%), Gaps = 1/39 (2%)
Frame = -2
Query: 79 GTGWQTTGRKGGGSGKGGKGGGGGYRIPIPGV-GKGGGG 116
G G TG GG G G+GGGG PG+ G+GGGG
Sbjct: 196 GRGGA*TGAPPGGGGIPGRGGGG------PGIPGRGGGG 98
>TC8210 homologue to PIR|T10586|T10586 small nuclear
ribonucleoprotein-associated protein homolog
F9F13.90 - Arabidopsis thaliana {Arabidopsis
thaliana;}, partial (16%)
Length = 698
Score = 34.3 bits (77), Expect = 0.006
Identities = 18/30 (60%), Positives = 19/30 (63%)
Frame = -3
Query: 86 GRKGGGSGKGGKGGGGGYRIPIPGVGKGGG 115
GR GGG G G+GGGGG I PG GGG
Sbjct: 183 GRGGGGPGIPGRGGGGGPLIICPG---GGG 103
Score = 33.5 bits (75), Expect = 0.010
Identities = 19/39 (48%), Positives = 22/39 (55%), Gaps = 1/39 (2%)
Frame = -3
Query: 79 GTGWQTTGRKGGGSGKGGKGGGGGYRIPIPGV-GKGGGG 116
G G TG GG G G+GGGG PG+ G+GGGG
Sbjct: 234 GRGGA*TGAPPGGGGIPGRGGGG------PGIPGRGGGG 136
>AV426815
Length = 391
Score = 33.5 bits (75), Expect = 0.010
Identities = 16/28 (57%), Positives = 19/28 (67%), Gaps = 1/28 (3%)
Frame = +2
Query: 90 GGSGKGGKGGGGGYRIPI-PGVGKGGGG 116
GG +GG GGGGG+ + GVG GGGG
Sbjct: 104 GGDSEGGCGGGGGWEWEL*NGVGCGGGG 187
>AV410891
Length = 380
Score = 33.1 bits (74), Expect = 0.012
Identities = 13/29 (44%), Positives = 20/29 (68%)
Frame = -3
Query: 86 GRKGGGSGKGGKGGGGGYRIPIPGVGKGG 114
GR+GGG G+G GG+R+ + G+ +GG
Sbjct: 267 GRRGGGLAWTGRGRRGGHRLRLGGIERGG 181
>TC18628 weakly similar to PIR|T09527|T09527 glycine-rich protein 1 -
chickpea {Cicer arietinum;} , partial (32%)
Length = 424
Score = 33.1 bits (74), Expect = 0.012
Identities = 16/28 (57%), Positives = 17/28 (60%)
Frame = +3
Query: 89 GGGSGKGGKGGGGGYRIPIPGVGKGGGG 116
GGG G GG+GGGGG G GGGG
Sbjct: 9 GGGRGHGGRGGGGGR-------GGGGGG 71
Score = 30.4 bits (67), Expect = 0.081
Identities = 14/25 (56%), Positives = 17/25 (68%)
Frame = +3
Query: 79 GTGWQTTGRKGGGSGKGGKGGGGGY 103
G G + G +GGG G+G GGGGGY
Sbjct: 6 GGGGRGHGGRGGGGGRG--GGGGGY 74
>AV415539
Length = 419
Score = 33.1 bits (74), Expect = 0.012
Identities = 17/35 (48%), Positives = 19/35 (53%)
Frame = -2
Query: 82 WQTTGRKGGGSGKGGKGGGGGYRIPIPGVGKGGGG 116
W+ G GGG G GGGGG R G+GGGG
Sbjct: 163 WRGCGLPGGGRVGWGGGGGGGAR------GRGGGG 77
Score = 27.3 bits (59), Expect = 0.69
Identities = 10/14 (71%), Positives = 11/14 (78%)
Frame = -2
Query: 89 GGGSGKGGKGGGGG 102
GGG G G+GGGGG
Sbjct: 115 GGGGGARGRGGGGG 74
Score = 24.3 bits (51), Expect = 5.8
Identities = 11/21 (52%), Positives = 12/21 (56%)
Frame = -2
Query: 81 GWQTTGRKGGGSGKGGKGGGG 101
GW G GGG +G GGGG
Sbjct: 127 GW---GGGGGGGARGRGGGGG 74
>AV773560
Length = 438
Score = 32.7 bits (73), Expect = 0.016
Identities = 15/24 (62%), Positives = 16/24 (66%)
Frame = +3
Query: 86 GRKGGGSGKGGKGGGGGYRIPIPG 109
GR GGG G G+GGGGG I PG
Sbjct: 363 GRGGGGPGIPGRGGGGGPLIICPG 434
Score = 27.3 bits (59), Expect = 0.69
Identities = 18/37 (48%), Positives = 18/37 (48%)
Frame = +3
Query: 79 GTGWQTTGRKGGGSGKGGKGGGGGYRIPIPGVGKGGG 115
G G TG GG GG G GG IPG G GGG
Sbjct: 312 GGGGA*TGAPAGG---GGIPGRGGGGPGIPGRGGGGG 413
>TC15523 similar to UP|GRPA_MAIZE (P10979) Glycine-rich RNA-binding,
abscisic acid-inducible protein, complete
Length = 972
Score = 32.7 bits (73), Expect = 0.016
Identities = 20/42 (47%), Positives = 21/42 (49%), Gaps = 8/42 (19%)
Frame = +2
Query: 83 QTTGRKGGGSG--------KGGKGGGGGYRIPIPGVGKGGGG 116
Q GR GGG G +GG GGGGG R G GGGG
Sbjct: 587 QARGRGGGGGGGGYGGGRREGGYGGGGGSRYS----GGGGGG 700
Score = 31.6 bits (70), Expect = 0.036
Identities = 17/38 (44%), Positives = 19/38 (49%)
Frame = +2
Query: 79 GTGWQTTGRKGGGSGKGGKGGGGGYRIPIPGVGKGGGG 116
G G + G GGG + GGGGGY G GGGG
Sbjct: 629 GGGRREGGYGGGGGSRYSGGGGGGYG------GSGGGG 724
Score = 31.2 bits (69), Expect = 0.047
Identities = 18/38 (47%), Positives = 20/38 (52%)
Frame = +2
Query: 79 GTGWQTTGRKGGGSGKGGKGGGGGYRIPIPGVGKGGGG 116
G G G GGG G +GGGGGY G +GGGG
Sbjct: 686 GGGGGYGGSGGGGYGGRREGGGGGY----GGSREGGGG 787
Score = 30.8 bits (68), Expect = 0.062
Identities = 16/36 (44%), Positives = 18/36 (49%)
Frame = +2
Query: 81 GWQTTGRKGGGSGKGGKGGGGGYRIPIPGVGKGGGG 116
G T + G+GG GGGGGY G GGGG
Sbjct: 560 GRNVTVNEAQARGRGGGGGGGGYGGGRREGGYGGGG 667
Score = 29.3 bits (64), Expect = 0.18
Identities = 15/28 (53%), Positives = 15/28 (53%)
Frame = +2
Query: 89 GGGSGKGGKGGGGGYRIPIPGVGKGGGG 116
GGG G G GGGGY G G G GG
Sbjct: 683 GGGGGGYGGSGGGGYGGRREGGGGGYGG 766
Score = 26.2 bits (56), Expect = 1.5
Identities = 14/27 (51%), Positives = 16/27 (58%), Gaps = 1/27 (3%)
Frame = +2
Query: 79 GTGWQTTGRKGGGSGKGG-KGGGGGYR 104
G G R+GGG G GG + GGGG R
Sbjct: 713 GGGGYGGRREGGGGGYGGSREGGGGSR 793
Score = 25.8 bits (55), Expect = 2.0
Identities = 17/36 (47%), Positives = 19/36 (52%)
Frame = +2
Query: 79 GTGWQTTGRKGGGSGKGGKGGGGGYRIPIPGVGKGG 114
G G + +G GGG G G GGG G R G G GG
Sbjct: 662 GGGSRYSGGGGGGYG-GSGGGGYGGRREGGGGGYGG 766
Score = 25.4 bits (54), Expect = 2.6
Identities = 13/27 (48%), Positives = 14/27 (51%)
Frame = +2
Query: 91 GSGKGGKGGGGGYRIPIPGVGKGGGGK 117
G G GG GGG G G G GGG +
Sbjct: 596 GRGGGGGGGGYGGGRREGGYGGGGGSR 676
>TC14068 homologue to UP|Q9SLR7 (Q9SLR7) Thiamin biosynthetic enzyme,
partial (85%)
Length = 1494
Score = 32.3 bits (72), Expect = 0.021
Identities = 13/25 (52%), Positives = 16/25 (64%)
Frame = -2
Query: 78 KGTGWQTTGRKGGGSGKGGKGGGGG 102
+G GW + GG G GG+GGGGG
Sbjct: 125 RGWGWDFGVEE*GGGGGGGEGGGGG 51
>TC10777 similar to UP|Q9SCL3 (Q9SCL3) Pre-mRNA splicing factor SF2-like
protein, partial (46%)
Length = 597
Score = 32.0 bits (71), Expect = 0.028
Identities = 19/62 (30%), Positives = 28/62 (44%)
Frame = +1
Query: 41 LKGNEGKSNDGCGDGIEIHCRKGYVDGTIWDKDISITKGTGWQTTGRKGGGSGKGGKGGG 100
++G +G + DGC +E+ G G ++ R+G G G GG GGG
Sbjct: 421 IRGRDGYNFDGCRLRVEL-----------------AHGGRGPTSSDRRGYGGGGGGGGGG 549
Query: 101 GG 102
GG
Sbjct: 550 GG 555
>TC19911 weakly similar to PIR|A44805|A44805 eggshell protein precursor -
fluke (Schistosoma haematobium) {Schistosoma
haematobium;} , partial (24%)
Length = 350
Score = 32.0 bits (71), Expect = 0.028
Identities = 27/88 (30%), Positives = 35/88 (39%), Gaps = 3/88 (3%)
Frame = +1
Query: 32 NTNECGGAILKGNEGKSNDGCGDGIEIHCRKGYVDGTIWDKDISI--TKGTGWQTTGRKG 89
NT GG GN+ + +D G + G +D D G G+ G
Sbjct: 28 NTGHSGGGS-HGNDHRHSDNEHGGRRSETTGHGLSGGGYDDDNRSGGRTGGGYGDGGHSA 204
Query: 90 GGSGK-GGKGGGGGYRIPIPGVGKGGGG 116
GG G GG+ GGGG G+ G GG
Sbjct: 205 GGYGDHGGRSGGGGGGYGDGGLSSGSGG 288
Database: LJGI
Posted date: Jul 30, 2004 11:16 AM
Number of letters in database: 14,692,800
Number of sequences in database: 28,460
Lambda K H
0.311 0.143 0.454
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 2,272,522
Number of Sequences: 28460
Number of extensions: 38280
Number of successful extensions: 1221
Number of sequences better than 10.0: 342
Number of HSP's better than 10.0 without gapping: 724
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 970
length of query: 117
length of database: 4,897,600
effective HSP length: 78
effective length of query: 39
effective length of database: 2,677,720
effective search space: 104431080
effective search space used: 104431080
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.7 bits)
S2: 49 (23.5 bits)
Medicago: description of AC138451.8


