

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC137824.11 - phase: 0
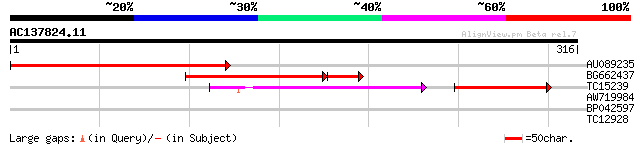
(316 letters)
Database: LJGI
28,460 sequences; 14,692,800 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
AU089235 192 5e-50
BG662437 156 6e-45
TC15239 UP|Q852R1 (Q852R1) Serine palmitoyltransferase, partial ... 64 3e-19
AW719984 30 0.41
BP042597 27 3.5
TC12928 26 7.7
>AU089235
Length = 407
Score = 192 bits (489), Expect = 5e-50
Identities = 92/123 (74%), Positives = 106/123 (85%)
Frame = +1
Query: 1 MASSFINFVNTTIDLVTYALHAPSARAVVFGFNIGGHLFIEVLLLVVILFLLSQKSYKPP 60
MA++ ++FVNT + + +A+ APSARAVVFG NIGGHLF+EV LLVVILFLLSQKSYKPP
Sbjct: 19 MAATVVDFVNTALGWLRFAVDAPSARAVVFGVNIGGHLFLEVFLLVVILFLLSQKSYKPP 198
Query: 61 KRPLSNKEIDELCDEWVPQPLIPSLNDEMPYEPPVLESAAGPHTIVNGKEVVNFASANYL 120
KRPL+NKEIDELCDEW P LIPSLN+ M YEPP L SAAGPHTI NGK+VVNF+S NYL
Sbjct: 199 KRPLTNKEIDELCDEWAPXSLIPSLNEXMLYEPPXLXSAAGPHTIXNGKDVVNFSSXNYL 378
Query: 121 GLI 123
GL+
Sbjct: 379 GLV 387
>BG662437
Length = 313
Score = 156 bits (395), Expect(2) = 6e-45
Identities = 72/79 (91%), Positives = 77/79 (97%)
Frame = +1
Query: 99 AAGPHTIVNGKEVVNFASANYLGLIGHQKLLDSCSSALEKYGVGSCGPRGFYGTIDVHLD 158
AAGPHTIVNGK+VVNF+SANYLGL+GHQKLLDSC+SALEKYGVGSCGPRGFYGTIDVHLD
Sbjct: 16 AAGPHTIVNGKDVVNFSSANYLGLVGHQKLLDSCTSALEKYGVGSCGPRGFYGTIDVHLD 195
Query: 159 CEGRISKFLGTPDSILYSY 177
CE RI+KFLGTPD ILYSY
Sbjct: 196 CEARIAKFLGTPD*ILYSY 252
Score = 40.8 bits (94), Expect(2) = 6e-45
Identities = 19/20 (95%), Positives = 20/20 (100%)
Frame = +2
Query: 178 GLSTMFSAIPAFSKKGDIIV 197
GLSTM+SAIPAFSKKGDIIV
Sbjct: 254 GLSTMYSAIPAFSKKGDIIV 313
>TC15239 UP|Q852R1 (Q852R1) Serine palmitoyltransferase, partial (81%)
Length = 1896
Score = 63.5 bits (153), Expect(2) = 3e-19
Identities = 39/126 (30%), Positives = 62/126 (48%), Gaps = 5/126 (3%)
Frame = +1
Query: 112 VNFASANYLGLIGHQ-----KLLDSCSSALEKYGVGSCGPRGFYGTIDVHLDCEGRISKF 166
+N S NYLG +++DS L+KY +C R GT D+H + E ++ F
Sbjct: 28 LNLGSYNYLGFAASDEYCTPRVIDS----LKKYSPSTCSARVDGGTTDLHNELEECVASF 195
Query: 167 LGTPDSILYSYGLSTMFSAIPAFSKKGDIIVADEGVHWGIQNGLYLSRSTVVYFKHNNMD 226
+ P ++++ G T + +P KG +I++D H I NG S +T+ F+HN
Sbjct: 196 VRKPAALVFGMGYVTNSAILPVLMGKGSLIISDSLNHNSIVNGARGSGATIRVFQHNMPS 375
Query: 227 SLRETL 232
L E L
Sbjct: 376 HLEEVL 393
Score = 47.4 bits (111), Expect(2) = 3e-19
Identities = 21/54 (38%), Positives = 34/54 (62%)
Frame = +2
Query: 249 IVIEALYQNSGQIAPLDEIIKLKEKYCFRILLDESNSFGVLGSSGRGLTEHYGV 302
+++E +Y G++ L EII + +KY LDE++S G +G +GRG+ E GV
Sbjct: 452 VIVEGIYSMEGELCKLPEIIAICKKYKAYTYLDEAHSIGAVGKTGRGVRELLGV 613
>AW719984
Length = 437
Score = 30.4 bits (67), Expect = 0.41
Identities = 16/66 (24%), Positives = 32/66 (48%)
Frame = +2
Query: 109 KEVVNFASANYLGLIGHQKLLDSCSSALEKYGVGSCGPRGFYGTIDVHLDCEGRISKFLG 168
K+++ F+ +YLGL H + + + A +++G+G G G + H E ++
Sbjct: 233 KKLILFSGNDYLGLSSHPTIGRAAAKAAQEHGMGPRGSALICGYTNYHRLXESSLADLKN 412
Query: 169 TPDSIL 174
D +L
Sbjct: 413 KEDCLL 430
>BP042597
Length = 414
Score = 27.3 bits (59), Expect = 3.5
Identities = 12/41 (29%), Positives = 21/41 (50%)
Frame = -2
Query: 70 DELCDEWVPQPLIPSLNDEMPYEPPVLESAAGPHTIVNGKE 110
+E C W P + + + + PP L A P+T++ G+E
Sbjct: 410 EENCCRWAKDVDTPCVCELLVHLPPFLVKPAHPYTLIVGEE 288
>TC12928
Length = 755
Score = 26.2 bits (56), Expect = 7.7
Identities = 16/47 (34%), Positives = 26/47 (55%), Gaps = 1/47 (2%)
Frame = -1
Query: 32 FNIGGHLFIEVLLLVVILFL-LSQKSYKPPKRPLSNKEIDELCDEWV 77
+++G H I V+ I+ L L Q PK P ++ +I LC+EW+
Sbjct: 200 WHLGSHFQIFVIDRS*IMSLPLLQIKQVHPKCPCTSIDIKPLCNEWI 60
Database: LJGI
Posted date: Jul 30, 2004 11:16 AM
Number of letters in database: 14,692,800
Number of sequences in database: 28,460
Lambda K H
0.321 0.141 0.422
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 5,759,168
Number of Sequences: 28460
Number of extensions: 85984
Number of successful extensions: 389
Number of sequences better than 10.0: 12
Number of HSP's better than 10.0 without gapping: 387
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 389
length of query: 316
length of database: 4,897,600
effective HSP length: 90
effective length of query: 226
effective length of database: 2,336,200
effective search space: 527981200
effective search space used: 527981200
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 55 (25.8 bits)
Medicago: description of AC137824.11


