

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC136449.7 - phase: 0
(471 letters)
Database: LJGI
28,460 sequences; 14,692,800 total letters
Searching..................................................done
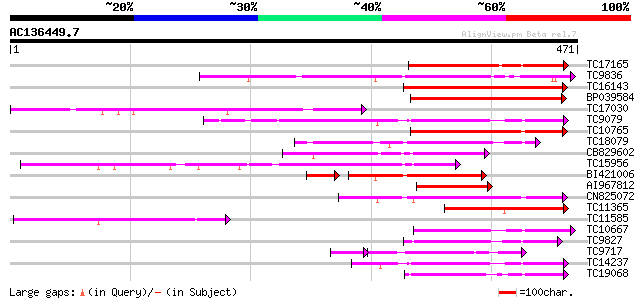
Score E
Sequences producing significant alignments: (bits) Value
TC17165 similar to UP|Q9SMG6 (Q9SMG6) Betanidin-5-O-glucosyltran... 171 2e-43
TC9836 weakly similar to UP|Q9LXV0 (Q9LXV0) Glucosyltransferase-... 169 7e-43
TC16143 similar to UP|Q7XZD0 (Q7XZD0) Isoflavonoid glucosyltrans... 156 8e-39
BP039584 155 1e-38
TC17030 similar to UP|Q8S995 (Q8S995) Glucosyltransferase-14, pa... 147 3e-36
TC9079 weakly similar to UP|Q8W4G1 (Q8W4G1) UDP-glucose glucosyl... 142 9e-35
TC10765 homologue to UP|Q9ZWQ5 (Q9ZWQ5) UDP-glycose:flavonoid gl... 130 5e-31
TC18079 weakly similar to UP|UFO5_MANES (Q40287) Flavonol 3-O-gl... 126 7e-30
CB829602 125 2e-29
TC15956 weakly similar to UP|LGT_CITUN (Q9MB73) Limonoid UDP-glu... 117 4e-27
BI421006 111 4e-27
AI967812 110 4e-25
CN825072 109 1e-24
TC11365 similar to UP|Q8S995 (Q8S995) Glucosyltransferase-14, pa... 106 7e-24
TC11585 similar to UP|Q9ZWQ5 (Q9ZWQ5) UDP-glycose:flavonoid glyc... 104 4e-23
TC10667 similar to UP|Q9M051 (Q9M051) Glucuronosyl transferase-l... 98 3e-21
TC9827 weakly similar to UP|Q8S9A2 (Q8S9A2) Glucosyltransferase-... 97 7e-21
TC9717 weakly similar to UP|Q9LNI1 (Q9LNI1) F6F3.22 protein, par... 83 5e-19
TC14237 weakly similar to UP|AAR06913 (AAR06913) UDP-glycosyltra... 90 7e-19
TC19068 weakly similar to UP|AAR06913 (AAR06913) UDP-glycosyltra... 89 2e-18
>TC17165 similar to UP|Q9SMG6 (Q9SMG6) Betanidin-5-O-glucosyltransferase,
partial (24%)
Length = 546
Score = 171 bits (433), Expect = 2e-43
Identities = 84/133 (63%), Positives = 106/133 (79%)
Frame = +1
Query: 332 IFRGWAPQLLILEHEAVGGFMTHCGWNSTLEGVCAGVPMATWPLSAEQFINEKLITDVLR 391
I RGWAPQ+LILEHEA+G F+THCGWNSTLEGV AGVPM TWP++AEQF NEKL+T+VL+
Sbjct: 1 IIRGWAPQVLILEHEAIGAFVTHCGWNSTLEGVAAGVPMVTWPIAAEQFYNEKLVTEVLK 180
Query: 392 IGVQVGSREWGSWDEERKELVGREKVELAVKKLMAESEDTEEMRRRVKSIFENAKRAVEE 451
IGV VG+++WG E + V + VE VK++M E E+ EEMR RVK + + A+ AVEE
Sbjct: 181 IGVPVGAKKWGRMVLE-GDGVKSDAVEKGVKRIM-EGEEAEEMRNRVKVLSQQAQWAVEE 354
Query: 452 GGSSYDDIHALIQ 464
GGSSY D++ALI+
Sbjct: 355 GGSSYSDLNALIE 393
>TC9836 weakly similar to UP|Q9LXV0 (Q9LXV0) Glucosyltransferase-like
protein, partial (55%)
Length = 1286
Score = 169 bits (429), Expect = 7e-43
Identities = 115/327 (35%), Positives = 177/327 (53%), Gaps = 14/327 (4%)
Frame = +1
Query: 158 DSEPFIVPGLPDIIEMTRSQTPIFMRNPS-QFSDRIKQLEE-----NSLGTLINSFYDLE 211
DS+ F +P P+ ++ R+Q P + + + + Q+ NS G L N+ D +
Sbjct: 79 DSDHFPLPDFPEAGQIHRTQLPSNIAEADGEDAWSLFQISNISNWVNSDGFLFNTVADFD 258
Query: 212 PAYADYVRNKLGKKAWLVGPVSLCNRSVEDKKERGKQPTIDEQSCLNWLNSKKPNSVLYI 271
YV KL + AW +GPV L S RGK I + C WL++K NSVL++
Sbjct: 259 SMGLRYVARKLNRPAWAIGPVLLSTGS----GSRGKGGGISPELCRKWLDTKPSNSVLFV 426
Query: 272 SFGSVARVPMKQLKEIAYGLEASDQSFIWVV----GKILNSSKNEEDWVLDKFERRMKEM 327
SFGS+ + Q+ ++A L+ S ++FIWVV G +NS E+W+ + F R+ +
Sbjct: 427 SFGSMNTISASQMMQLATALDRSGRNFIWVVRPPIGFDINSEFRAEEWLPEGFLGRVAK- 603
Query: 328 DKGLIFRGWAPQLLILEHEAVGGFMTHCGWNSTLEGVCAGVPMATWPLSAEQFINEKLIT 387
KGL+ WAPQ+ IL H AV F++HCGWNS LE + GVP+ WP++AEQF N K++
Sbjct: 604 -KGLVVHDWAPQVEILSHGAVSAFLSHCGWNSVLESLSHGVPILGWPMAAEQFFNCKMLE 780
Query: 388 DVLRIGVQVGSREWGSWDEERKELVGREKVELAVKKLMAESEDTEEMRRRVKSIFENAKR 447
+ L + V+V + + ++LV EK+EL +M E+E ++R+ +I E +
Sbjct: 781 EELGVCVEVARGK--RCEVRHEDLV--EKIEL----VMNEAESGVKIRKNAGNIREMIRD 936
Query: 448 AV--EEG--GSSYDDIHALIQCKTSLD 470
AV E+G GSS I + SL+
Sbjct: 937 AVRDEDGYKGSSVRAIDEFLSAAMSLN 1017
>TC16143 similar to UP|Q7XZD0 (Q7XZD0) Isoflavonoid glucosyltransferase,
partial (30%)
Length = 604
Score = 156 bits (394), Expect = 8e-39
Identities = 76/137 (55%), Positives = 103/137 (74%), Gaps = 1/137 (0%)
Frame = +1
Query: 328 DKGLIFRGWAPQLLILEHEAVGGFMTHCGWNSTLEGVCAGVPMATWPLSAEQFINEKLIT 387
+KG+I RGW PQ++IL H AVG F+THCGWNST+E V AGVPM TWP+ EQF NEKLIT
Sbjct: 4 EKGMIIRGWVPQVVILGHRAVGAFVTHCGWNSTVEAVSAGVPMITWPVVGEQFYNEKLIT 183
Query: 388 DVLRIGVQVGSREWGSWD-EERKELVGREKVELAVKKLMAESEDTEEMRRRVKSIFENAK 446
+V IGV+VG+ E ++R++LV RE +E AV++LM ++ E++RRR + E A+
Sbjct: 184 EVRGIGVEVGAEECCLMGFDKREKLVSRESIEKAVRRLMDGGDEGEQIRRRAQEYGEKAR 363
Query: 447 RAVEEGGSSYDDIHALI 463
+AVEEGGSS+ ++ ALI
Sbjct: 364 QAVEEGGSSHKNLTALI 414
>BP039584
Length = 545
Score = 155 bits (393), Expect = 1e-38
Identities = 71/129 (55%), Positives = 94/129 (72%)
Frame = -3
Query: 334 RGWAPQLLILEHEAVGGFMTHCGWNSTLEGVCAGVPMATWPLSAEQFINEKLITDVLRIG 393
RGWAPQLLILEH A+GG +THCGWN+TLE V AG+PMAT PL AEQF NEKL+ DVL +G
Sbjct: 543 RGWAPQLLILEHPAIGGVVTHCGWNTTLESVIAGLPMATMPLFAEQFYNEKLLVDVLGVG 364
Query: 394 VQVGSREWGSWDEERKELVGREKVELAVKKLMAESEDTEEMRRRVKSIFENAKRAVEEGG 453
V +G ++W +W+ E+V RE + A+ LM E+ EMRRRV+ + + AK+ ++ GG
Sbjct: 363 VSIGMKKWRNWNVVGDEIVKRENIVKAISLLMGGGEEALEMRRRVRELSDAAKKTIQPGG 184
Query: 454 SSYDDIHAL 462
SSY+ + L
Sbjct: 183 SSYNKVKGL 157
>TC17030 similar to UP|Q8S995 (Q8S995) Glucosyltransferase-14, partial (51%)
Length = 1098
Score = 147 bits (372), Expect = 3e-36
Identities = 105/315 (33%), Positives = 151/315 (47%), Gaps = 19/315 (6%)
Frame = +1
Query: 1 MNHETDAVKIYFFPFVGGGHQIPMVDTARVFAKH-GATSTIITTPSNALQFQNSITRDQK 59
M + + FP + GH IPM+D A + ++ T T+ITTP NA +F T K
Sbjct: 187 MASQASQIHFVLFPLMSQGHMIPMMDIATILSQQQDVTVTVITTPHNASRF----THTTK 354
Query: 60 SNHPITIQILTTPENT-------EVTDTDMSAGPMID----TSILLEPLKQFL--VQHRP 106
S I I L P N E D S G + S L +P++Q + P
Sbjct: 355 SRSQIRILELEFPHNETGLPEGCENLDMLPSLGTALTFFNAASNLQKPVEQLFNELTPPP 534
Query: 107 DGIVVDMFHRWAGDIIDELKIPRIVFNGNGCFPRCVIENTRKHVVLENLSSDSEPFIVPG 166
+ I+ DM + I + IPRI F G CF + N +H V +N++S++E F++PG
Sbjct: 535 NCIISDMCLPYTARIAAKFNIPRISFLGQSCFTLFCLYNIGRHKVRQNVTSETEYFVLPG 714
Query: 167 LPDIIEMTRSQTP-----IFMRNPSQFSDRIKQLEENSLGTLINSFYDLEPAYADYVRNK 221
+PD IEMT++Q P +N F E S G ++NSF +LE YA +
Sbjct: 715 VPDKIEMTKAQIPGPSGARMDQNRIAFYAETAAAEAASYGVVMNSFEELETEYAKGYKKV 894
Query: 222 LGKKAWLVGPVSLCNRSVEDKKERGKQPTIDEQSCLNWLNSKKPNSVLYISFGSVARVPM 281
K W +GPVSL N + E DE C+ WL+ +K SV+Y GS+ +
Sbjct: 895 KNGKVWCIGPVSLSNNNKVSIDE-------DEHYCMKWLDLQKSKSVIYACLGSMCNLTP 1053
Query: 282 KQLKEIAYGLEASDQ 296
QL E+ GLEAS++
Sbjct: 1054LQLIELGLGLEASNR 1098
>TC9079 weakly similar to UP|Q8W4G1 (Q8W4G1) UDP-glucose
glucosyltransferase, partial (65%)
Length = 1585
Score = 142 bits (359), Expect = 9e-35
Identities = 97/307 (31%), Positives = 155/307 (49%), Gaps = 4/307 (1%)
Frame = +1
Query: 162 FIVPGLPDIIEMTRSQTPIFMRNPSQFSDRIKQLEENSLGTLINSFYDLEPAYADYVRNK 221
F + LPD I T I + + +DR K+ + N+F +LE +
Sbjct: 574 FRLKDLPDFIRTT-DPNDIMLEFMVEVADRAKRAS----AIVFNTFNELERDVLSALSTM 738
Query: 222 LGKKAWLVGPV-SLCNRSVEDK-KERGKQPTIDEQSCLNWLNSKKPNSVLYISFGSVARV 279
L + +GP S N++ +++ G ++ CL WL SK+P SV+Y++FGS+ +
Sbjct: 739 L-PSLYPIGPFPSFLNQAPQNQLASLGSNLWKEDTKCLQWLESKEPGSVVYVNFGSITVM 915
Query: 280 PMKQLKEIAYGLEASDQSFIWVVGK--ILNSSKNEEDWVLDKFERRMKEMDKGLIFRGWA 337
+QL E A+GL + F+W++ + + S +D+ R GLI W
Sbjct: 916 SPEQLLEFAWGLANGKKPFLWIIRPDLVADGSAILSPKFVDETSNR------GLI-ASWC 1074
Query: 338 PQLLILEHEAVGGFMTHCGWNSTLEGVCAGVPMATWPLSAEQFINEKLITDVLRIGVQVG 397
PQ +L H +VGGF+THCGWNST+E +CAGVPM WP A+Q N + I + IG++V
Sbjct: 1075PQEKVLNHPSVGGFLTHCGWNSTIESICAGVPMLCWPFVADQPTNCRYICNEWDIGIEVD 1254
Query: 398 SREWGSWDEERKELVGREKVELAVKKLMAESEDTEEMRRRVKSIFENAKRAVEEGGSSYD 457
+ V RE+VE + +LM + ++MR+R + + A+ GG SY
Sbjct: 1255TN------------VKREEVENLIHELMV-GDKGKKMRQRTMELKKKAEEDTRPGGCSYM 1395
Query: 458 DIHALIQ 464
++ LI+
Sbjct: 1396NLDKLIK 1416
>TC10765 homologue to UP|Q9ZWQ5 (Q9ZWQ5) UDP-glycose:flavonoid
glycosyltransferase, partial (19%)
Length = 542
Score = 130 bits (327), Expect = 5e-31
Identities = 65/131 (49%), Positives = 92/131 (69%), Gaps = 1/131 (0%)
Frame = +2
Query: 334 RGWAPQLLILEHEAVGGFMTHCGWNSTLEGVCAGVPMATWPLSAEQFINEKLITDVLRIG 393
+GWAPQ LIL H A GGF+THCGWN+ +E + AGVPM T P ++Q+ NEKLIT+V G
Sbjct: 2 KGWAPQPLILNHLATGGFLTHCGWNAVVEAISAGVPMITMPGFSDQYYNEKLITEVHGFG 181
Query: 394 VQVGSREWG-SWDEERKELVGREKVELAVKKLMAESEDTEEMRRRVKSIFENAKRAVEEG 452
V+VG+ EW S + +K++V E++E AVK LM E+ E+R + K + + A +AV++G
Sbjct: 182 VEVGAAEWSISPYDGKKKVVSGERIEKAVKSLM--GEEGAEIRSKAKEVQDKAWKAVQQG 355
Query: 453 GSSYDDIHALI 463
GSSY+ + LI
Sbjct: 356 GSSYNSLTVLI 388
>TC18079 weakly similar to UP|UFO5_MANES (Q40287) Flavonol
3-O-glucosyltransferase 5 (UDP-glucose flavonoid
3-O-glucosyltransferase 5) , partial (33%)
Length = 889
Score = 126 bits (317), Expect = 7e-30
Identities = 81/220 (36%), Positives = 116/220 (51%), Gaps = 15/220 (6%)
Frame = +3
Query: 237 RSVEDKKERGKQPTIDEQSCLNWLNSKKPNSVLYISFGSVARVPMKQLKEIAYGLEASDQ 296
R+VE K G+ D+ L WL+ + SV+Y+SFGS +P Q+ EIA GLE S
Sbjct: 15 RTVESKPRHGE----DQDEILRWLDKQPAGSVIYVSFGSGGTMPQGQMTEIALGLELSQH 182
Query: 297 SFIWVVGKILNSSKNEED---------------WVLDKFERRMKEMDKGLIFRGWAPQLL 341
F+WVV NE D ++ + F R +E G++ WAPQ
Sbjct: 183 RFVWVV-----RPPNEADASATFFGFGNEMALNYLPEGFMNRTREF--GVVVPMWAPQAE 341
Query: 342 ILEHEAVGGFMTHCGWNSTLEGVCAGVPMATWPLSAEQFINEKLITDVLRIGVQVGSREW 401
IL H A GGF+T CGWNS LE V GVPM WPL +EQ +N ++++ L + V+V E
Sbjct: 342 ILGHPATGGFVTPCGWNSVLESVLNGVPMVAWPLYSEQKMNAYMLSEELGVAVRVKVAEG 521
Query: 402 GSWDEERKELVGREKVELAVKKLMAESEDTEEMRRRVKSI 441
G +V R++V V+++M SE+ M+ RV+ +
Sbjct: 522 G--------VVCRDQVVDMVRRVMV-SEEGMGMKDRVREL 614
>CB829602
Length = 542
Score = 125 bits (313), Expect = 2e-29
Identities = 64/174 (36%), Positives = 104/174 (58%), Gaps = 2/174 (1%)
Frame = +1
Query: 227 WLVGPVSLCNRSVEDKKERGKQPTI--DEQSCLNWLNSKKPNSVLYISFGSVARVPMKQL 284
+ +GP+ L V + + + + +E CL WL+S++P+SVLY++FGSV + +QL
Sbjct: 1 YTIGPLDLLLNQVTENRIESIKCNLWKEEPECLKWLDSQEPSSVLYVNFGSVINMTPQQL 180
Query: 285 KEIAYGLEASDQSFIWVVGKILNSSKNEEDWVLDKFERRMKEMDKGLIFRGWAPQLLILE 344
E+A+G+ S + FIWV+ L + E VL + K D+G++ W PQ +L+
Sbjct: 181 VELAWGIANSKKKFIWVIRPDL--VEGEASIVLPEIVAETK--DRGIML-SWCPQEQVLK 345
Query: 345 HEAVGGFMTHCGWNSTLEGVCAGVPMATWPLSAEQFINEKLITDVLRIGVQVGS 398
H A+GGF+THCGWNST+E + +GVP+ P +QF+N + I G+++ S
Sbjct: 346 HSALGGFLTHCGWNSTIESISSGVPLICSPFFNDQFVNSRYICSEWDFGMEMNS 507
>TC15956 weakly similar to UP|LGT_CITUN (Q9MB73) Limonoid
UDP-glucosyltransferase (Limonoid glucosyltransferase)
(Limonoid GTase) (LGTase) , partial (65%)
Length = 1169
Score = 117 bits (293), Expect = 4e-27
Identities = 99/390 (25%), Positives = 177/390 (45%), Gaps = 25/390 (6%)
Frame = +2
Query: 10 IYFFPFVGGGHQIPMVDTARVFAKHGATSTIITTPSNALQFQNSITRDQKSNHPITIQIL 69
I F GH P++ A+ A G++ T+ TT + + + K+ PI+ L
Sbjct: 41 ILLVSFPAQGHINPLLRLAKCLAAKGSSVTLATTQTAGKSMRTATNITDKAPTPISDGSL 220
Query: 70 TTP--ENTEVTDTDMSAG---------PMIDTSILLEPLKQFLVQHRPDGIVVDM-FHRW 117
++ D D G ++ +L+ +K+ +R +++ F W
Sbjct: 221 KFEFFDDGLGPDDDSIRGNLSLYLQQLELVGRQQILKMIKEHADSNRQISCIINNPFLPW 400
Query: 118 AGDIIDELKIPRIVF--NGNGCFPRCVIENTRKHVVLENL---SSDSEPFIVPGLPDI-I 171
D+ +E IP + + F T + NL S +P LP I
Sbjct: 401 VVDVAEEQGIPSALLWIQSSAVF-------TAYYSYFHNLVTFPSKEDPLADVQLPSTSI 559
Query: 172 EMTRSQTPIFMRNPSQFS-------DRIKQLEENSLGTLINSFYDLEPAYADYVRNKLGK 224
+ + P F+ + ++ K L + + L++++ +LE + DY+
Sbjct: 560 VLKHCEIPDFLHPSCTYPFLGTLILEQFKNLNK-TFCILVDTYEELEHDFIDYL-----S 721
Query: 225 KAWLVGPVSLCNRSVEDKKERGKQPTIDEQSCLNWLNSKKPNSVLYISFGSVARVPMKQL 284
K +L+ PV +S + K + + C+ WLNSK +SV+Y+SFGS+ +P +Q+
Sbjct: 722 KTFLIRPVGPLFKSSQAKSAAIRGDFMKSDDCIEWLNSKPQSSVVYVSFGSIVYLPQEQV 901
Query: 285 KEIAYGLEASDQSFIWVVGKILNSSKNEEDWVLDKFERRMKEMDKGLIFRGWAPQLLILE 344
EIA+GL+ S SF+WV+ + + + F E +G + + W+PQ +L
Sbjct: 902 DEIAFGLKKSQVSFLWVMKPPPEVTGLQPHVLPYGFLEETGE--RGRVVQ-WSPQEEVLA 1072
Query: 345 HEAVGGFMTHCGWNSTLEGVCAGVPMATWP 374
H +V F+THCGWNS++E + +GVP+ T+P
Sbjct: 1073HPSVSCFLTHCGWNSSMEALTSGVPVLTFP 1162
>BI421006
Length = 519
Score = 111 bits (277), Expect(2) = 4e-27
Identities = 56/120 (46%), Positives = 80/120 (66%), Gaps = 5/120 (4%)
Frame = +2
Query: 282 KQLKEIAYGLEASDQSFIWVV-----GKILNSSKNEEDWVLDKFERRMKEMDKGLIFRGW 336
+Q+++IA GLE S Q FIWV+ G I + K +E + FE+R++ M GL+ R W
Sbjct: 128 EQIEQIANGLEQSKQKFIWVLRDADKGDIFDGDKVKERGLPKGFEKRVEGM--GLVVRDW 301
Query: 337 APQLLILEHEAVGGFMTHCGWNSTLEGVCAGVPMATWPLSAEQFINEKLITDVLRIGVQV 396
APQL IL H + GGFM+HCGWNS +E + GVP+A WP+ ++Q N LIT+VL++ + V
Sbjct: 302 APQLEILSHPSTGGFMSHCGWNSCIESMSMGVPIAAWPMHSDQPRNTVLITEVLKVALVV 481
Score = 26.9 bits (58), Expect(2) = 4e-27
Identities = 10/28 (35%), Positives = 17/28 (60%)
Frame = +3
Query: 247 KQPTIDEQSCLNWLNSKKPNSVLYISFG 274
K ++ + WL+ ++ SVLY+SFG
Sbjct: 24 KNSSMGRNFVMEWLDRQEQKSVLYVSFG 107
>AI967812
Length = 274
Score = 110 bits (276), Expect = 4e-25
Identities = 49/63 (77%), Positives = 56/63 (88%)
Frame = +2
Query: 339 QLLILEHEAVGGFMTHCGWNSTLEGVCAGVPMATWPLSAEQFINEKLITDVLRIGVQVGS 398
Q+LILEHEA+G F+THCGWNSTLEGV AGVPM TWPLSAEQF NEKL+T VL+IGV VG
Sbjct: 17 QVLILEHEAIGAFVTHCGWNSTLEGVSAGVPMVTWPLSAEQFYNEKLVTHVLKIGVPVGV 196
Query: 399 REW 401
++W
Sbjct: 197 KKW 205
>CN825072
Length = 712
Score = 109 bits (272), Expect = 1e-24
Identities = 68/200 (34%), Positives = 104/200 (52%), Gaps = 10/200 (5%)
Frame = -3
Query: 274 GSVARVPMKQLKEIAYGLEASDQSFIWVVGK--ILNSSKNEEDWVLDKFERRMKEMDKGL 331
GS Q+ EIA +E S F+W + K + S D+ +D + E G
Sbjct: 710 GSRGSFDRAQITEIARAVENSGVRFVWSLRKPXLKGSMVGPSDYSVDDLASVLPE---GF 540
Query: 332 IFR--------GWAPQLLILEHEAVGGFMTHCGWNSTLEGVCAGVPMATWPLSAEQFINE 383
+ R GWAPQ +L H A GGF++HCGWNSTLE + GVP+ATWPL AEQ N
Sbjct: 539 LDRTTGIGRVIGWAPQTRVLTHPATGGFVSHCGWNSTLESIYFGVPIATWPLYAEQQTNA 360
Query: 384 KLITDVLRIGVQVGSREWGSWDEERKELVGREKVELAVKKLMAESEDTEEMRRRVKSIFE 443
++ L+I V++ + L+ +K+E ++ ++ + E+R+RVK + E
Sbjct: 359 FVLVRELKIAVEISLDYRVEINGGPNYLLTADKIEGGIRSVL---DKDGEVRKRVKEMSE 189
Query: 444 NAKRAVEEGGSSYDDIHALI 463
+++ + EGG SY + LI
Sbjct: 188 KSRKTLLEGGCSYSYLDRLI 129
>TC11365 similar to UP|Q8S995 (Q8S995) Glucosyltransferase-14, partial (24%)
Length = 498
Score = 106 bits (265), Expect = 7e-24
Identities = 55/105 (52%), Positives = 72/105 (68%), Gaps = 2/105 (1%)
Frame = +3
Query: 362 EGVCAGVPMATWPLSAEQFINEKLITDVLRIGVQVGSREWGSWDEERK--ELVGREKVEL 419
E +C GVPM TWPL +QF +EKLI +L +GV+VG W EE + LV +E VE
Sbjct: 3 EAICGGVPMVTWPLFGDQFFHEKLIVQILGVGVRVGVEAPVKWGEEEEIGVLVKKEDVER 182
Query: 420 AVKKLMAESEDTEEMRRRVKSIFENAKRAVEEGGSSYDDIHALIQ 464
A+++LM ES + EMRRRV+ + E AKRAVEEGGSS+ ++ LIQ
Sbjct: 183 AIEELMNESSPSHEMRRRVQELAEMAKRAVEEGGSSHTNVTLLIQ 317
>TC11585 similar to UP|Q9ZWQ5 (Q9ZWQ5) UDP-glycose:flavonoid
glycosyltransferase, partial (40%)
Length = 609
Score = 104 bits (259), Expect = 4e-23
Identities = 62/191 (32%), Positives = 97/191 (50%), Gaps = 11/191 (5%)
Frame = +1
Query: 4 ETDAVKIYFFPFVGGGHQIPMVDTARVFAKHGATSTIITTPSNALQFQNSITRDQKSNHP 63
E + +K+YF PF GH IP+V AR+ A G TIITTP+NA F +I D+ +
Sbjct: 16 EMETLKVYFVPFFAQGHLIPLVHLARLVASRGEHVTIITTPANAQLFDKTIEEDRACGYH 195
Query: 64 ITIQILTTP----------ENTEVTDTDMSAGPMIDTSILLEP-LKQFLVQHRPDGIVVD 112
I + ++ P EN +++AG + + L++P ++ F+ Q PD ++ D
Sbjct: 196 IRVHLVKFPSTQVGLPGGVENLFAASDNLTAGKIHMAAHLIQPEIEGFMKQSPPDVLIPD 375
Query: 113 MFHRWAGDIIDELKIPRIVFNGNGCFPRCVIENTRKHVVLENLSSDSEPFIVPGLPDIIE 172
+ W+ L IPR++FN F C+IE + H SDS P+ +PGLP +
Sbjct: 376 IMFTWSEASAKSLGIPRLIFNPISIFDVCMIEAIKSHPEAFT-CSDSGPYHIPGLPHSLT 552
Query: 173 MTRSQTPIFMR 183
+ +P F R
Sbjct: 553 LPIKPSPGFAR 585
>TC10667 similar to UP|Q9M051 (Q9M051) Glucuronosyl transferase-like
protein, partial (20%)
Length = 824
Score = 98.2 bits (243), Expect = 3e-21
Identities = 52/135 (38%), Positives = 76/135 (55%)
Frame = +1
Query: 336 WAPQLLILEHEAVGGFMTHCGWNSTLEGVCAGVPMATWPLSAEQFINEKLITDVLRIGVQ 395
WAPQ +L+H A+G F THCGWNSTLE VC GVPM P +Q +N K ++DV ++GVQ
Sbjct: 22 WAPQEQVLKHPAIGAFWTHCGWNSTLESVCEGVPMICSPCFGDQKVNAKYVSDVWKVGVQ 201
Query: 396 VGSREWGSWDEERKELVGREKVELAVKKLMAESEDTEEMRRRVKSIFENAKRAVEEGGSS 455
+ ++ GR ++E + LM ++ E+R + + E A + EGGSS
Sbjct: 202 LQNKP------------GRGEIEKTISLLML-GDEAYEIRGNILKLKEKANVCLSEGGSS 342
Query: 456 YDDIHALIQCKTSLD 470
Y + L+ SL+
Sbjct: 343 YCFLDRLVSEILSLE 387
>TC9827 weakly similar to UP|Q8S9A2 (Q8S9A2) Glucosyltransferase-7
(Fragment), partial (42%)
Length = 611
Score = 96.7 bits (239), Expect = 7e-21
Identities = 55/132 (41%), Positives = 79/132 (59%)
Frame = +2
Query: 328 DKGLIFRGWAPQLLILEHEAVGGFMTHCGWNSTLEGVCAGVPMATWPLSAEQFINEKLIT 387
+KGLI W PQLL+L+H+A+G F+THCGWNSTLE V GVP+ PL + N KLI
Sbjct: 20 EKGLIVT-WCPQLLVLKHQALGCFLTHCGWNSTLESVSLGVPVIAMPLWTDPVTNGKLIV 196
Query: 388 DVLRIGVQVGSREWGSWDEERKELVGREKVELAVKKLMAESEDTEEMRRRVKSIFENAKR 447
DV + GV+ + E KE+V RE ++ +K+++ E+E E++ AK
Sbjct: 197 DVWKTGVRAVADE--------KEIVRRETIKNCIKEII-ETEKINEIKSNAIKWMNLAKD 349
Query: 448 AVEEGGSSYDDI 459
++ EGG S +I
Sbjct: 350 SLAEGGRSDKNI 385
>TC9717 weakly similar to UP|Q9LNI1 (Q9LNI1) F6F3.22 protein, partial (18%)
Length = 902
Score = 82.8 bits (203), Expect(2) = 5e-19
Identities = 42/132 (31%), Positives = 73/132 (54%)
Frame = +2
Query: 298 FIWVVGKILNSSKNEEDWVLDKFERRMKEMDKGLIFRGWAPQLLILEHEAVGGFMTHCGW 357
F WV+ K + S + DW+ ++ + KG+++R WAPQ+ IL H+++GGF+THCGW
Sbjct: 98 FFWVLKKQNSDSADSHDWIENQSNK------KGMVWRTWAPQMRILAHKSIGGFLTHCGW 259
Query: 358 NSTLEGVCAGVPMATWPLSAEQFINEKLITDVLRIGVQVGSREWGSWDEERKELVGREKV 417
+S +EG+ G P+ P EQ + + + + ++GV+V + R+ V
Sbjct: 260 SSVIEGLQVGCPLIMLPFPNEQMLVARFM-EGKKLGVKVSK-------NQHDGKFTRDSV 415
Query: 418 ELAVKKLMAESE 429
A++ +M E E
Sbjct: 416 AKALRSVMLEEE 451
Score = 28.1 bits (61), Expect(2) = 5e-19
Identities = 13/32 (40%), Positives = 19/32 (58%)
Frame = +1
Query: 267 SVLYISFGSVARVPMKQLKEIAYGLEASDQSF 298
SV+Y++FGS + + E+A GLE S F
Sbjct: 4 SVIYVAFGSEVTLSNEDFTELALGLELSGFPF 99
>TC14237 weakly similar to UP|AAR06913 (AAR06913) UDP-glycosyltransferase
85A8, partial (27%)
Length = 999
Score = 90.1 bits (222), Expect = 7e-19
Identities = 60/184 (32%), Positives = 92/184 (49%), Gaps = 4/184 (2%)
Frame = +2
Query: 285 KEI-AYGLEASDQSFIWVVGKIL---NSSKNEEDWVLDKFERRMKEMDKGLIFRGWAPQL 340
KEI +G+ S F+W++ + S N LD+ + R G I W Q
Sbjct: 176 KEIWLWGIANSKVPFLWILRPDVVMGEDSINVPQEFLDEIKGR------GYI-ASWCFQE 334
Query: 341 LILEHEAVGGFMTHCGWNSTLEGVCAGVPMATWPLSAEQFINEKLITDVLRIGVQVGSRE 400
+L H ++G F+THCGWNST+EG+ AG+P+ WP +EQ N + IG++V
Sbjct: 335 EVLSHPSIGAFLTHCGWNSTIEGISAGLPLICWPFFSEQHTNSRYACTTWEIGMEVNHD- 511
Query: 401 WGSWDEERKELVGREKVELAVKKLMAESEDTEEMRRRVKSIFENAKRAVEEGGSSYDDIH 460
V R+++ V ++M + E +EMR++ + A A GGSSY+D H
Sbjct: 512 -----------VKRDEITTLVNEMM-KGEKGKEMRKKGLEWKKKAIEATGLGGSSYNDFH 655
Query: 461 ALIQ 464
LI+
Sbjct: 656 KLIK 667
>TC19068 weakly similar to UP|AAR06913 (AAR06913) UDP-glycosyltransferase
85A8, partial (26%)
Length = 565
Score = 88.6 bits (218), Expect = 2e-18
Identities = 50/136 (36%), Positives = 76/136 (55%)
Frame = +1
Query: 329 KGLIFRGWAPQLLILEHEAVGGFMTHCGWNSTLEGVCAGVPMATWPLSAEQFINEKLITD 388
+GLI W Q +L H ++GGF+THCGWNST+E + AGVPM WP A+Q N + I +
Sbjct: 49 RGLI-ASWCSQEHVLNHPSIGGFLTHCGWNSTIESISAGVPMLCWPFFADQLTNCRSICN 225
Query: 389 VLRIGVQVGSREWGSWDEERKELVGREKVELAVKKLMAESEDTEEMRRRVKSIFENAKRA 448
IG+Q+ D K RE+VE + +LM E ++MR++ + + A+
Sbjct: 226 EWDIGMQI--------DTNGK----REEVEKLINELMV-GEKGKKMRQKTMELKKRAEED 366
Query: 449 VEEGGSSYDDIHALIQ 464
GG SY ++ +I+
Sbjct: 367 TRPGGCSYMNLDQVIK 414
Database: LJGI
Posted date: Jul 30, 2004 11:16 AM
Number of letters in database: 14,692,800
Number of sequences in database: 28,460
Lambda K H
0.318 0.136 0.406
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 7,924,040
Number of Sequences: 28460
Number of extensions: 104070
Number of successful extensions: 586
Number of sequences better than 10.0: 99
Number of HSP's better than 10.0 without gapping: 555
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 555
length of query: 471
length of database: 4,897,600
effective HSP length: 94
effective length of query: 377
effective length of database: 2,222,360
effective search space: 837829720
effective search space used: 837829720
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 57 (26.6 bits)
Medicago: description of AC136449.7


