

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC136138.6 - phase: 0 /pseudo
(339 letters)
Database: LJGI
28,460 sequences; 14,692,800 total letters
Searching..................................................done
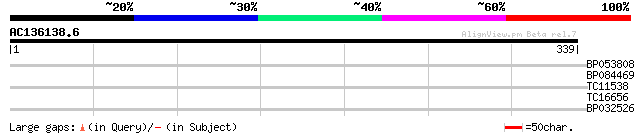
Score E
Sequences producing significant alignments: (bits) Value
BP053808 30 0.58
BP084469 28 2.2
TC11538 28 2.9
TC16656 UP|RR19_LOTJA (Q9BBP7) Chloroplast 30S ribosomal protein... 27 3.8
BP032526 27 4.9
>BP053808
Length = 462
Score = 30.0 bits (66), Expect = 0.58
Identities = 13/29 (44%), Positives = 18/29 (61%), Gaps = 1/29 (3%)
Frame = +2
Query: 186 WLRDELGSTIFIQQ-DNARTHINPNDPEF 213
W D + TI+ + +T+INPNDPEF
Sbjct: 86 WDTDNIPYTIYFENASKEKTNINPNDPEF 172
>BP084469
Length = 485
Score = 28.1 bits (61), Expect = 2.2
Identities = 11/23 (47%), Positives = 15/23 (64%)
Frame = -1
Query: 251 HYKEAPKTIDELVNAVVKSFENY 273
HY+E P+ I ELV V + F +Y
Sbjct: 341 HYRELPQEIQELVGPVPEGFNDY 273
>TC11538
Length = 542
Score = 27.7 bits (60), Expect = 2.9
Identities = 11/26 (42%), Positives = 16/26 (61%)
Frame = +1
Query: 54 LSMLEGIPHDPMFKSMYNIIHIDEKW 79
L+ + G PH P+ K +Y IIH+ W
Sbjct: 256 LNSVPGTPHLPLGKILYRIIHLLVDW 333
>TC16656 UP|RR19_LOTJA (Q9BBP7) Chloroplast 30S ribosomal protein S19,
complete
Length = 473
Score = 27.3 bits (59), Expect = 3.8
Identities = 8/22 (36%), Positives = 16/22 (72%)
Frame = -2
Query: 59 GIPHDPMFKSMYNIIHIDEKWF 80
G+P+DP+ KS+ + H + +W+
Sbjct: 205 GLPYDPLHKSVNALFHCE*QWY 140
>BP032526
Length = 516
Score = 26.9 bits (58), Expect = 4.9
Identities = 17/55 (30%), Positives = 25/55 (44%), Gaps = 1/55 (1%)
Frame = +1
Query: 153 VAGTMETKAITSINRDLIRSFLIEKVLPATKEVWLRD-ELGSTIFIQQDNARTHI 206
+AG KA + RDL R + +PAT V R+ E G+ + + R I
Sbjct: 178 IAGIRLPKAANELRRDLRRGIKVRVAIPATGPVHSREREPGAAVRVLHGKQRRKI 342
Database: LJGI
Posted date: Jul 30, 2004 11:16 AM
Number of letters in database: 14,692,800
Number of sequences in database: 28,460
Lambda K H
0.327 0.140 0.415
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 5,234,225
Number of Sequences: 28460
Number of extensions: 67091
Number of successful extensions: 403
Number of sequences better than 10.0: 10
Number of HSP's better than 10.0 without gapping: 402
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 403
length of query: 339
length of database: 4,897,600
effective HSP length: 91
effective length of query: 248
effective length of database: 2,307,740
effective search space: 572319520
effective search space used: 572319520
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 15 ( 7.1 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.7 bits)
S2: 55 (25.8 bits)
Medicago: description of AC136138.6


