

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC135161.4 + phase: 0
(467 letters)
Database: LJGI
28,460 sequences; 14,692,800 total letters
Searching..................................................done
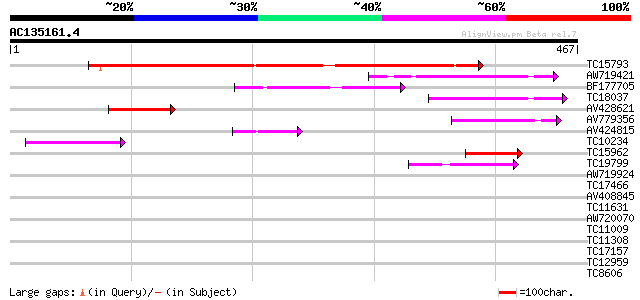
Score E
Sequences producing significant alignments: (bits) Value
TC15793 similar to UP|Q9FGH9 (Q9FGH9) Leucine zipper protein (AT... 323 5e-89
AW719421 111 3e-25
BF177705 108 2e-24
TC18037 weakly similar to UP|Q9FGH9 (Q9FGH9) Leucine zipper prot... 86 2e-17
AV428621 66 1e-11
AV779356 53 1e-07
AV424815 51 5e-07
TC10234 49 2e-06
TC15962 similar to UP|Q9FHC6 (Q9FHC6) Emb|CAB83315.1, partial (13%) 45 3e-05
TC19799 weakly similar to PIR|PQ0476|PQ0476 pistil extensin-like... 42 2e-04
AW719924 33 0.13
TC17466 similar to UP|Q9M427 (Q9M427) Oligouridylate binding pro... 32 0.17
AV408845 32 0.17
TC11631 weakly similar to UP|TCH2_ARATH (P25070) Calmodulin-rela... 32 0.29
AW720070 32 0.29
TC11009 similar to GB|AAP21377.1|30102918|BT006569 At1g47970 {Ar... 31 0.49
TC11308 similar to UP|CENB_HUMAN (P07199) Major centromere autoa... 31 0.49
TC17157 similar to UP|CAD27462 (CAD27462) Nucleosome assembly pr... 30 0.64
TC12959 similar to UP|AAS52042 (AAS52042) ADR122Cp, partial (6%) 30 0.64
TC8606 similar to UP|AAQ82688 (AAQ82688) Epa4p, partial (7%) 30 0.64
>TC15793 similar to UP|Q9FGH9 (Q9FGH9) Leucine zipper protein
(AT5g58430/mqj2_20), partial (43%)
Length = 969
Score = 323 bits (827), Expect = 5e-89
Identities = 176/328 (53%), Positives = 226/328 (68%), Gaps = 3/328 (0%)
Frame = +2
Query: 66 LTIKDVNM---EDLGLRIKRWIKAFKVAFKILFPTERQLCDIVFFEFSAISDISFTDVCR 122
L+I+DV+ +DL I+RW+KA KVA ILFP+ER+LCD VFF FS+ +D+SF +VCR
Sbjct: 17 LSIEDVHKLPWQDLEDEIERWMKASKVALMILFPSERRLCDRVFFGFSSAADLSFMEVCR 196
Query: 123 EFTIRLLNFPNVIANDQSNTTLLFRMLDMYETLHDLIPNFESLFCDQYSVSLRNELNTVL 182
I+LLNF + +A + LFR+LD+YET+ DLIP FESLFCDQYSVSLRNE T+
Sbjct: 197 GTAIQLLNFADAVAIGTRSPERLFRVLDVYETMRDLIPEFESLFCDQYSVSLRNEAITIW 376
Query: 183 KKLGETIVGTLREFENTIRSKGPGNAPFFGGQLHPLVRFVMNFLTWICDYREILEQVFED 242
K+LGE I G E EN IR + P A GG LHP+ R+VMN+L C + LEQVFED
Sbjct: 377 KRLGEAIRGVFMELENLIR-RDPAKAEVPGGGLHPITRYVMNYLRAACRSLQTLEQVFED 553
Query: 243 HGHVLLEYTKHDDTVPSSSSSSSSSSSSSSSLQMERIMEVLESKLEAMFNIFNDPTLGYV 302
+GH L EY + DD VP SSSS S+QM+ IME+LES LEA I+ DP L YV
Sbjct: 554 YGHPLKEYPRLDDRVP---------SSSSMSVQMDWIMELLESNLEAKSKIYKDPALCYV 706
Query: 303 YLMNSSRYIIIKTMENELGTLLGDGMLQRHSAKLRYNFEEYIRSSWGKVLEFLRLDNNLL 362
+LMN+ RYI+ KT ++ELGTLLGD +++H+AK+R Y RSSW KVL FL++D +
Sbjct: 707 FLMNNGRYIVQKTRDSELGTLLGDDWIRKHTAKVRQYHVLYQRSSWNKVLGFLKMDTGSM 886
Query: 363 VHPNMVGKSMKKQLKSFNKLFNEICKAQ 390
V KSMK++L+SFN LF +IC+ Q
Sbjct: 887 PSGG-VAKSMKEKLRSFNILFEDICRVQ 967
>AW719421
Length = 575
Score = 111 bits (277), Expect = 3e-25
Identities = 65/157 (41%), Positives = 93/157 (58%)
Frame = +2
Query: 296 DPTLGYVYLMNSSRYIIIKTMENELGTLLGDGMLQRHSAKLRYNFEEYIRSSWGKVLEFL 355
DP L Y ++MN+ RYI +++L T LGD Q+ K + NFE Y RSSW VLEFL
Sbjct: 5 DPALCYFFMMNNMRYI-----DSQLXTTLGDDRFQK---KAQQNFELYCRSSWSNVLEFL 160
Query: 356 RLDNNLLVHPNMVGKSMKKQLKSFNKLFNEICKAQSLWFIMDETLKEEIIVYLGENLLPA 415
+LD + N+ KSM ++L F + F E+C QS W + D+ L+++II + LLPA
Sbjct: 161 KLDIDESWDSNVAAKSMIEKLNLFYQHFKEVCGVQSTWRVCDKQLRKQIIESVENMLLPA 340
Query: 416 YTNFIRKLHIVLKLEVKKPPDGYIEYETKDIKAILNN 452
Y NFI + VL + D +I+YE DI+ +LN+
Sbjct: 341 YRNFIEQFKDVL----GRQSDEFIKYEMCDIQGMLNS 439
>BF177705
Length = 412
Score = 108 bits (269), Expect = 2e-24
Identities = 63/141 (44%), Positives = 81/141 (56%)
Frame = +2
Query: 186 GETIVGTLREFENTIRSKGPGNAPFFGGQLHPLVRFVMNFLTWICDYREILEQVFEDHGH 245
G+ I E EN IR G +HP+ VMN+L C RE LE VF+++GH
Sbjct: 14 GKAITWIFMELENLIRDSAKTEVS--DGGVHPITCDVMNYLRAACRSRETLEHVFKEYGH 187
Query: 246 VLLEYTKHDDTVPSSSSSSSSSSSSSSSLQMERIMEVLESKLEAMFNIFNDPTLGYVYLM 305
L EY + DD VPSSSS +Q+ IME+LES LEA I+ DP LG V+LM
Sbjct: 188 PLKEYPEFDDKVPSSSSMP---------VQVNLIMELLESSLEAKSKIYKDPALGDVFLM 340
Query: 306 NSSRYIIIKTMENELGTLLGD 326
N+ RYI+ + + ELGTLLG+
Sbjct: 341 NNVRYIVQEAKDGELGTLLGN 403
>TC18037 weakly similar to UP|Q9FGH9 (Q9FGH9) Leucine zipper protein
(AT5g58430/mqj2_20), partial (8%)
Length = 495
Score = 85.5 bits (210), Expect = 2e-17
Identities = 48/114 (42%), Positives = 67/114 (58%)
Frame = +1
Query: 346 SSWGKVLEFLRLDNNLLVHPNMVGKSMKKQLKSFNKLFNEICKAQSLWFIMDETLKEEII 405
SSW K+L+ L++++N PN+ MK +L FN F EIC+ QS W ++DE L+EEI
Sbjct: 4 SSWNKMLDILKVESNESEAPNVAADLMKDKLNLFNMHFEEICRVQSTWSVIDEQLREEIR 183
Query: 406 VYLGENLLPAYTNFIRKLHIVLKLEVKKPPDGYIEYETKDIKAILNNMFKIYRP 459
+ + + LLPAY NFI + VL K YIEY DI+ LN +F +P
Sbjct: 184 ISVNDILLPAYGNFIGRFQDVL----GKHAYEYIEYGLFDIEDSLNYLFVGAKP 333
>AV428621
Length = 230
Score = 66.2 bits (160), Expect = 1e-11
Identities = 31/55 (56%), Positives = 39/55 (70%)
Frame = +2
Query: 82 RWIKAFKVAFKILFPTERQLCDIVFFEFSAISDISFTDVCREFTIRLLNFPNVIA 136
RW+KA VA KILFP ER LCD + FS+ +D SFT+ CR I+LLNF +V+A
Sbjct: 53 RWVKASNVALKILFPNER*LCDRILLGFSSAADFSFTEDCRGSVIQLLNFADVVA 217
>AV779356
Length = 518
Score = 52.8 bits (125), Expect = 1e-07
Identities = 34/90 (37%), Positives = 46/90 (50%)
Frame = -3
Query: 365 PNMVGKSMKKQLKSFNKLFNEICKAQSLWFIMDETLKEEIIVYLGENLLPAYTNFIRKLH 424
P + MK++L FN ++ + QS W + D L+EEI L LLPAY FI +
Sbjct: 363 PMLKQNLMKEKLDLFNFCLEQMSRVQSTWRVYDNKLREEIRTSLENVLLPAYGIFIGRFP 184
Query: 425 IVLKLEVKKPPDGYIEYETKDIKAILNNMF 454
VL K YIEY DI+ LN++F
Sbjct: 183 DVLGKHAHK----YIEYGMFDIQERLNHLF 106
Score = 31.6 bits (70), Expect = 0.29
Identities = 13/39 (33%), Positives = 27/39 (68%)
Frame = -2
Query: 339 NFEEYIRSSWGKVLEFLRLDNNLLVHPNMVGKSMKKQLK 377
N E Y+RSSW KV + L+L+++ L+ ++ +S +++ +
Sbjct: 442 NLELYLRSSWDKVFDLLKLNHHELMVSDVEAESNERKAR 326
>AV424815
Length = 309
Score = 50.8 bits (120), Expect = 5e-07
Identities = 24/58 (41%), Positives = 35/58 (59%)
Frame = +2
Query: 184 KLGETIVGTLREFENTIRSKGPGNAPFFGGQLHPLVRFVMNFLTWICDYREILEQVFE 241
+LGE GT EFEN IR++ P G +HPL+R+VMN+L + DY ++ + E
Sbjct: 2 RLGEAAKGTFTEFENCIRNE-TSKKPVLTGDVHPLLRYVMNYLKLLVDYGAAMDSLLE 172
>TC10234
Length = 902
Score = 48.5 bits (114), Expect = 2e-06
Identities = 27/83 (32%), Positives = 48/83 (57%), Gaps = 1/83 (1%)
Frame = +1
Query: 14 EALSLETVNNLQETVKLMLNSGFNKECLIVYSSCRRECLEECLVKQFLNSDNLT-IKDVN 72
E +S+ + +L+ + M+ G+ KEC+ VY+ R+ ++E L + NL ++ ++
Sbjct: 637 ERVSMLAMEDLKAIAESMIACGYGKECVKVYTIMRKSIVDEALYHLGVERLNLPQVQKMD 816
Query: 73 MEDLGLRIKRWIKAFKVAFKILF 95
E L L+I+ W+KA KVA LF
Sbjct: 817 WEVLELKIRSWLKAVKVAVGTLF 885
>TC15962 similar to UP|Q9FHC6 (Q9FHC6) Emb|CAB83315.1, partial (13%)
Length = 602
Score = 44.7 bits (104), Expect = 3e-05
Identities = 19/47 (40%), Positives = 31/47 (65%)
Frame = +3
Query: 376 LKSFNKLFNEICKAQSLWFIMDETLKEEIIVYLGENLLPAYTNFIRK 422
LK FN +F E+ + QS W + D L+E + + + E LLPAY +F+++
Sbjct: 3 LKQFNVMFEELHQKQSQWTVPDSELRESLRLAVAEVLLPAYRSFVKR 143
>TC19799 weakly similar to PIR|PQ0476|PQ0476 pistil extensin-like protein
(clone pMG08) - common tobacco
(fragment) {Nicotiana tabacum;} , partial (12%)
Length = 601
Score = 42.4 bits (98), Expect = 2e-04
Identities = 27/92 (29%), Positives = 44/92 (47%), Gaps = 1/92 (1%)
Frame = +2
Query: 329 LQRHSAKLRYNFEEYIRSSWGKVLEFLRLDNNLLVHPNMVGK-SMKKQLKSFNKLFNEIC 387
L H K++ +Y R +W KV+ L +P G+ + + +FN F C
Sbjct: 8 LANHKLKVKEYVSKYERVAWNKVVAALP------ENPTAAGEVTARACFVAFNAAFR*EC 169
Query: 388 KAQSLWFIMDETLKEEIIVYLGENLLPAYTNF 419
+ QS W + D L+EEI +G L+ Y++F
Sbjct: 170 RKQSSWVVSDPKLREEIKGSIGSMLVLKYSDF 265
>AW719924
Length = 505
Score = 32.7 bits (73), Expect = 0.13
Identities = 18/28 (64%), Positives = 21/28 (74%)
Frame = +1
Query: 258 PSSSSSSSSSSSSSSSLQMERIMEVLES 285
PSSSSSSSSSSSSSS Q+ + + ES
Sbjct: 10 PSSSSSSSSSSSSSSHFQLPPPLPINES 93
>TC17466 similar to UP|Q9M427 (Q9M427) Oligouridylate binding protein,
partial (20%)
Length = 389
Score = 32.3 bits (72), Expect = 0.17
Identities = 17/20 (85%), Positives = 19/20 (95%)
Frame = +3
Query: 256 TVPSSSSSSSSSSSSSSSLQ 275
+VP SSSSSSSSSSSSS+LQ
Sbjct: 12 SVPLSSSSSSSSSSSSSALQ 71
>AV408845
Length = 429
Score = 32.3 bits (72), Expect = 0.17
Identities = 17/18 (94%), Positives = 17/18 (94%)
Frame = +3
Query: 258 PSSSSSSSSSSSSSSSLQ 275
PSSSSSSSSSSSSSSS Q
Sbjct: 93 PSSSSSSSSSSSSSSSFQ 146
Score = 28.9 bits (63), Expect = 1.9
Identities = 15/24 (62%), Positives = 20/24 (82%)
Frame = +3
Query: 249 EYTKHDDTVPSSSSSSSSSSSSSS 272
+++K ++ SSSSSSSSSSSSSS
Sbjct: 69 DFSKPTNSPSSSSSSSSSSSSSSS 140
Score = 26.9 bits (58), Expect = 7.1
Identities = 14/20 (70%), Positives = 17/20 (85%)
Frame = +3
Query: 254 DDTVPSSSSSSSSSSSSSSS 273
D + P++S SSSSSSSSSSS
Sbjct: 69 DFSKPTNSPSSSSSSSSSSS 128
Score = 26.6 bits (57), Expect = 9.3
Identities = 17/37 (45%), Positives = 21/37 (55%)
Frame = +3
Query: 259 SSSSSSSSSSSSSSSLQMERIMEVLESKLEAMFNIFN 295
SSSSSSSSSSSS L + S L ++ IF+
Sbjct: 105 SSSSSSSSSSSSFQPLTLFSFFTPQPSSLNSLCFIFS 215
>TC11631 weakly similar to UP|TCH2_ARATH (P25070) Calmodulin-related protein
2, touch-induced, partial (68%)
Length = 530
Score = 31.6 bits (70), Expect = 0.29
Identities = 19/50 (38%), Positives = 27/50 (54%), Gaps = 6/50 (12%)
Frame = +3
Query: 167 CDQYSVSLR-----NELNTVLKKLGETI-VGTLREFENTIRSKGPGNAPF 210
CD Y + NEL++VLKKLGE +G R+ + + + G GN F
Sbjct: 330 CDLYDLDKNGLISANELHSVLKKLGEKCSLGDCRKMISNVDADGDGNVNF 479
>AW720070
Length = 484
Score = 31.6 bits (70), Expect = 0.29
Identities = 17/20 (85%), Positives = 18/20 (90%)
Frame = -3
Query: 258 PSSSSSSSSSSSSSSSLQME 277
P+SSSSSSSSSSSSSSL E
Sbjct: 155 PASSSSSSSSSSSSSSLLSE 96
Score = 28.5 bits (62), Expect = 2.4
Identities = 14/17 (82%), Positives = 16/17 (93%)
Frame = -3
Query: 257 VPSSSSSSSSSSSSSSS 273
+PSS +SSSSSSSSSSS
Sbjct: 167 IPSSPASSSSSSSSSSS 117
>TC11009 similar to GB|AAP21377.1|30102918|BT006569 At1g47970 {Arabidopsis
thaliana;}, partial (44%)
Length = 700
Score = 30.8 bits (68), Expect = 0.49
Identities = 16/18 (88%), Positives = 17/18 (93%)
Frame = -2
Query: 256 TVPSSSSSSSSSSSSSSS 273
+VP SSSSSSSSSSSSSS
Sbjct: 243 SVPKSSSSSSSSSSSSSS 190
Score = 28.5 bits (62), Expect = 2.4
Identities = 16/23 (69%), Positives = 18/23 (77%)
Frame = -2
Query: 251 TKHDDTVPSSSSSSSSSSSSSSS 273
T++ SSSSSSSSSSSSSSS
Sbjct: 252 TRYSVPKSSSSSSSSSSSSSSSS 184
Score = 28.1 bits (61), Expect = 3.2
Identities = 16/24 (66%), Positives = 18/24 (74%)
Frame = -2
Query: 250 YTKHDDTVPSSSSSSSSSSSSSSS 273
Y+ + SSSSSSSSSSSSSSS
Sbjct: 246 YSVPKSSSSSSSSSSSSSSSSSSS 175
Score = 26.9 bits (58), Expect = 7.1
Identities = 15/22 (68%), Positives = 16/22 (72%)
Frame = -2
Query: 252 KHDDTVPSSSSSSSSSSSSSSS 273
K + SSSSSSSSSSSSS S
Sbjct: 234 KSSSSSSSSSSSSSSSSSSSGS 169
Score = 26.9 bits (58), Expect = 7.1
Identities = 14/15 (93%), Positives = 14/15 (93%)
Frame = -2
Query: 256 TVPSSSSSSSSSSSS 270
T PSSSSSSSSSSSS
Sbjct: 144 TPPSSSSSSSSSSSS 100
>TC11308 similar to UP|CENB_HUMAN (P07199) Major centromere autoantigen B
(Centromere protein B) (CENP-B), partial (4%)
Length = 756
Score = 30.8 bits (68), Expect = 0.49
Identities = 16/18 (88%), Positives = 16/18 (88%)
Frame = +1
Query: 255 DTVPSSSSSSSSSSSSSS 272
DT P SSSSSSSSSSSSS
Sbjct: 220 DTSPGSSSSSSSSSSSSS 273
>TC17157 similar to UP|CAD27462 (CAD27462) Nucleosome assembly protein
1-like protein 3, partial (81%)
Length = 1298
Score = 30.4 bits (67), Expect = 0.64
Identities = 16/16 (100%), Positives = 16/16 (100%)
Frame = -3
Query: 258 PSSSSSSSSSSSSSSS 273
PSSSSSSSSSSSSSSS
Sbjct: 873 PSSSSSSSSSSSSSSS 826
Score = 29.3 bits (64), Expect = 1.4
Identities = 17/28 (60%), Positives = 20/28 (70%)
Frame = -3
Query: 246 VLLEYTKHDDTVPSSSSSSSSSSSSSSS 273
+LL + + SSSSSSSSSSSSSSS
Sbjct: 894 LLLLFPSPSSSSSSSSSSSSSSSSSSSS 811
Score = 28.1 bits (61), Expect = 3.2
Identities = 17/28 (60%), Positives = 18/28 (63%)
Frame = -3
Query: 259 SSSSSSSSSSSSSSSLQMERIMEVLESK 286
SSSSSSSSSSSSSSS + SK
Sbjct: 843 SSSSSSSSSSSSSSSSSPSSSSSSISSK 760
Score = 27.7 bits (60), Expect = 4.2
Identities = 15/15 (100%), Positives = 15/15 (100%)
Frame = -3
Query: 259 SSSSSSSSSSSSSSS 273
SSSSSSSSSSSSSSS
Sbjct: 852 SSSSSSSSSSSSSSS 808
Score = 27.7 bits (60), Expect = 4.2
Identities = 15/15 (100%), Positives = 15/15 (100%)
Frame = -3
Query: 259 SSSSSSSSSSSSSSS 273
SSSSSSSSSSSSSSS
Sbjct: 846 SSSSSSSSSSSSSSS 802
Score = 27.7 bits (60), Expect = 4.2
Identities = 15/15 (100%), Positives = 15/15 (100%)
Frame = -3
Query: 259 SSSSSSSSSSSSSSS 273
SSSSSSSSSSSSSSS
Sbjct: 849 SSSSSSSSSSSSSSS 805
Score = 26.9 bits (58), Expect = 7.1
Identities = 14/21 (66%), Positives = 17/21 (80%)
Frame = -3
Query: 259 SSSSSSSSSSSSSSSLQMERI 279
SSSSSSSS SSSSSS+ + +
Sbjct: 816 SSSSSSSSPSSSSSSISSKSL 754
>TC12959 similar to UP|AAS52042 (AAS52042) ADR122Cp, partial (6%)
Length = 517
Score = 30.4 bits (67), Expect = 0.64
Identities = 22/54 (40%), Positives = 31/54 (56%), Gaps = 2/54 (3%)
Frame = -3
Query: 240 FEDHGHVLLEYTKHDDTVPSSSSSSSSSSSSSSSLQM--ERIMEVLESKLEAMF 291
FE H ++ ++ SSSSSS SSSSSSSSL + +R + LE ++F
Sbjct: 221 FESHQD---HHSSSSSSLSSSSSSSESSSSSSSSLLLPSDRESQNLEEGQSSIF 69
Score = 28.5 bits (62), Expect = 2.4
Identities = 19/43 (44%), Positives = 22/43 (50%)
Frame = -3
Query: 247 LLEYTKHDDTVPSSSSSSSSSSSSSSSLQMERIMEVLESKLEA 289
L + H D SSSSS SSSSSSS S +L S E+
Sbjct: 230 LNNFESHQDHHSSSSSSLSSSSSSSESSSSSSSSLLLPSDRES 102
>TC8606 similar to UP|AAQ82688 (AAQ82688) Epa4p, partial (7%)
Length = 1008
Score = 30.4 bits (67), Expect = 0.64
Identities = 16/16 (100%), Positives = 16/16 (100%)
Frame = +1
Query: 258 PSSSSSSSSSSSSSSS 273
PSSSSSSSSSSSSSSS
Sbjct: 352 PSSSSSSSSSSSSSSS 399
Score = 28.5 bits (62), Expect = 2.4
Identities = 15/16 (93%), Positives = 15/16 (93%)
Frame = +1
Query: 258 PSSSSSSSSSSSSSSS 273
P SSSSSSSSSSSSSS
Sbjct: 349 PPSSSSSSSSSSSSSS 396
Score = 26.9 bits (58), Expect = 7.1
Identities = 14/18 (77%), Positives = 15/18 (82%)
Frame = +1
Query: 256 TVPSSSSSSSSSSSSSSS 273
+ PSSSSSS SSSSSSS
Sbjct: 322 STPSSSSSSPPSSSSSSS 375
Database: LJGI
Posted date: Jul 30, 2004 11:16 AM
Number of letters in database: 14,692,800
Number of sequences in database: 28,460
Lambda K H
0.322 0.138 0.400
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 8,272,310
Number of Sequences: 28460
Number of extensions: 131149
Number of successful extensions: 4039
Number of sequences better than 10.0: 135
Number of HSP's better than 10.0 without gapping: 1408
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 2772
length of query: 467
length of database: 4,897,600
effective HSP length: 94
effective length of query: 373
effective length of database: 2,222,360
effective search space: 828940280
effective search space used: 828940280
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 57 (26.6 bits)
Medicago: description of AC135161.4


