

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
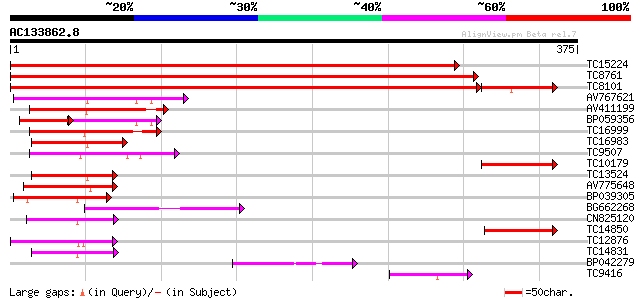
Query= AC133862.8 + phase: 0
(375 letters)
Database: LJGI
28,460 sequences; 14,692,800 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC15224 homologue to UP|Q9SWB5 (Q9SWB5) Seed maturation protein ... 568 e-163
TC8761 homologue to UP|Q9SWB5 (Q9SWB5) Seed maturation protein P... 561 e-161
TC8101 similar to UP|O24074 (O24074) DNAJ-like protein, partial ... 517 e-147
AV767621 100 5e-22
AV411199 91 2e-19
BP059356 56 3e-15
TC16999 similar to UP|Q9T024 (Q9T024) DNAJ-like protein (AT4g391... 72 2e-13
TC16983 similar to UP|Q9ZSY2 (Q9ZSY2) ALTERED response to gravit... 69 2e-12
TC9507 similar to UP|Q8VZB6 (Q8VZB6) DnaJ-like protein, partial ... 67 5e-12
TC10179 similar to UP|Q9SWB5 (Q9SWB5) Seed maturation protein PM... 65 2e-11
TC13524 similar to UP|O48678 (O48678) F3I6.4 protein, partial (19%) 64 3e-11
AV775648 63 7e-11
BP039305 57 5e-09
BG662268 56 1e-08
CN825120 55 2e-08
TC14850 homologue to UP|Q9SWB5 (Q9SWB5) Seed maturation protein ... 54 3e-08
TC12876 54 3e-08
TC14831 homologue to UP|P93499 (P93499) DnaJ-like protein (Fragm... 53 7e-08
BP042279 52 1e-07
TC9416 similar to UP|Q9FEW7 (Q9FEW7) DnaJ like protein, partial ... 51 4e-07
>TC15224 homologue to UP|Q9SWB5 (Q9SWB5) Seed maturation protein PM37,
partial (71%)
Length = 1031
Score = 568 bits (1465), Expect = e-163
Identities = 274/298 (91%), Positives = 285/298 (94%), Gaps = 1/298 (0%)
Frame = +3
Query: 1 MFGRAPKKSDNTKYYEILGVSKNASPDDLKKAYKKAAIKNHPDKGGDPEKFKELAQAYEV 60
MFGRAPKKSD+T+YYEILGV KNAS DDLKKAYKKAAIKNHPDKGGDPEKFKELAQAYEV
Sbjct: 138 MFGRAPKKSDSTRYYEILGVPKNASQDDLKKAYKKAAIKNHPDKGGDPEKFKELAQAYEV 317
Query: 61 LSDPEKREVYDQYGEDALKEGMGGGGGGHDPFDIFSSFFGGGGFP-GGGSSRGRRQRRGE 119
LSDPEKRE+YDQYGEDALKEGMGGGGGGHDPFDIF SFFGGGG P GGSSRGRRQRRGE
Sbjct: 318 LSDPEKREIYDQYGEDALKEGMGGGGGGHDPFDIFQSFFGGGGGPFSGGSSRGRRQRRGE 497
Query: 120 DVVHPLKVSLEDLYLGTSKKLSLSRNVLCSKCNGKGSKSGASMTCAGCQGSGMKISMRHL 179
DVVHPLKVSLEDLY GT+KKLSLSRNVLCSKCNGKGSKSGASM CAGCQG+GMK+S+RHL
Sbjct: 498 DVVHPLKVSLEDLYSGTAKKLSLSRNVLCSKCNGKGSKSGASMKCAGCQGTGMKVSIRHL 677
Query: 180 GANMIQQMQHPCNECKGTGETISDKDRCPQCKGEKVVQQKKVLEVHVEKGMQNGQKITFP 239
G +MIQQMQHPCNECKGTGETI+D+DRCPQCKGEKVVQ+KKVLEV VEKGMQNGQKITFP
Sbjct: 678 GPSMIQQMQHPCNECKGTGETINDRDRCPQCKGEKVVQEKKVLEVIVEKGMQNGQKITFP 857
Query: 240 GEADEAPDTVTGDIVFVLQQKEHPKFKRKGEDLFVEHTLSLTEALCGFQFALTHLDSR 297
GEADEAPDTVTGDIVFVLQQKEHPKFKRK EDLFVEHTLSLTEALCGFQF LTHLD R
Sbjct: 858 GEADEAPDTVTGDIVFVLQQKEHPKFKRKSEDLFVEHTLSLTEALCGFQFVLTHLDGR 1031
>TC8761 homologue to UP|Q9SWB5 (Q9SWB5) Seed maturation protein PM37,
partial (74%)
Length = 1050
Score = 561 bits (1447), Expect = e-161
Identities = 275/313 (87%), Positives = 291/313 (92%), Gaps = 3/313 (0%)
Frame = +3
Query: 1 MFGRAP-KKSDNTKYYEILGVSKNASPDDLKKAYKKAAIKNHPDKGGDPEKFKELAQAYE 59
MFGRAP KKSD+T+YY+ILGVSK AS DDLKKAYKKAAIKNHPDKGGDPEKFKELAQAYE
Sbjct: 78 MFGRAPPKKSDSTRYYDILGVSKTASQDDLKKAYKKAAIKNHPDKGGDPEKFKELAQAYE 257
Query: 60 VLSDPEKREVYDQYGEDALKEGMGGGGGG-HDPFDIFSSFFGGGGFP-GGGSSRGRRQRR 117
VL+DPEKRE+YD YGEDALKEGMGGGGGG HDPFDIFSSFFGG F GGG SRGRRQRR
Sbjct: 258 VLNDPEKREIYDNYGEDALKEGMGGGGGGGHDPFDIFSSFFGGSPFSSGGGGSRGRRQRR 437
Query: 118 GEDVVHPLKVSLEDLYLGTSKKLSLSRNVLCSKCNGKGSKSGASMTCAGCQGSGMKISMR 177
GEDVV+PLKVSLEDLYLG +KKLSLSRNVLCSKCNGKGSKSGAS+ C GCQGSGMK+S+R
Sbjct: 438 GEDVVYPLKVSLEDLYLGAAKKLSLSRNVLCSKCNGKGSKSGASLKCPGCQGSGMKVSIR 617
Query: 178 HLGANMIQQMQHPCNECKGTGETISDKDRCPQCKGEKVVQQKKVLEVHVEKGMQNGQKIT 237
H+G +MIQQMQHPCNECKGTGETI+D+DRCPQCKG+KV Q+KKVLEV VEKGMQN QKIT
Sbjct: 618 HIGPSMIQQMQHPCNECKGTGETINDRDRCPQCKGDKVSQEKKVLEVFVEKGMQNQQKIT 797
Query: 238 FPGEADEAPDTVTGDIVFVLQQKEHPKFKRKGEDLFVEHTLSLTEALCGFQFALTHLDSR 297
FPGEADEAPDT TGDIVFVLQ KEHPKFKRK EDLFVEHTLSLTEALCGFQF LTHLD R
Sbjct: 798 FPGEADEAPDTTTGDIVFVLQLKEHPKFKRKAEDLFVEHTLSLTEALCGFQFVLTHLDGR 977
Query: 298 QLLIKSNPGEVVK 310
QLLIKSNPGEVVK
Sbjct: 978 QLLIKSNPGEVVK 1016
>TC8101 similar to UP|O24074 (O24074) DNAJ-like protein, partial (90%)
Length = 1737
Score = 517 bits (1332), Expect = e-147
Identities = 239/312 (76%), Positives = 278/312 (88%)
Frame = +1
Query: 1 MFGRAPKKSDNTKYYEILGVSKNASPDDLKKAYKKAAIKNHPDKGGDPEKFKELAQAYEV 60
MFGRAP+KSDNTK+YE+LGV K+AS D++KKAY+KAA+KNHPDKGGDPEKFKEL QAYEV
Sbjct: 148 MFGRAPRKSDNTKFYEVLGVPKSASEDEIKKAYRKAAMKNHPDKGGDPEKFKELGQAYEV 327
Query: 61 LSDPEKREVYDQYGEDALKEGMGGGGGGHDPFDIFSSFFGGGGFPGGGSSRGRRQRRGED 120
LSDPEK+E+YDQYGEDALKEGMGGG H+PFDIF +FFGG GF GGGSSRGRRQR+GED
Sbjct: 328 LSDPEKKELYDQYGEDALKEGMGGGESFHNPFDIFETFFGGAGFGGGGSSRGRRQRQGED 507
Query: 121 VVHPLKVSLEDLYLGTSKKLSLSRNVLCSKCNGKGSKSGASMTCAGCQGSGMKISMRHLG 180
VVH +KVSLED+Y GT+KKLSLSRNVLC KC GKGSKSG + C GCQG+GMK+ R +G
Sbjct: 508 VVHSIKVSLEDVYNGTTKKLSLSRNVLCLKCKGKGSKSGTAGRCFGCQGTGMKVIRRQIG 687
Query: 181 ANMIQQMQHPCNECKGTGETISDKDRCPQCKGEKVVQQKKVLEVHVEKGMQNGQKITFPG 240
M+QQMQH C++C+GTGE IS++DRCPQCKG K+ Q+KKVLEVHVEKGM+ GQKI F G
Sbjct: 688 LGMVQQMQHVCSDCRGTGEVISERDRCPQCKGNKISQEKKVLEVHVEKGMRQGQKIVFEG 867
Query: 241 EADEAPDTVTGDIVFVLQQKEHPKFKRKGEDLFVEHTLSLTEALCGFQFALTHLDSRQLL 300
+ADEAPDT+TGDIVFV+Q KEHPKFKR+ +DL+++H LSLTEALCGFQFA+ HLD RQLL
Sbjct: 868 QADEAPDTITGDIVFVVQVKEHPKFKRELDDLYIDHNLSLTEALCGFQFAVKHLDGRQLL 1047
Query: 301 IKSNPGEVVKPG 312
IKSNPGEV+KPG
Sbjct: 1048IKSNPGEVIKPG 1083
Score = 40.0 bits (92), Expect = 6e-04
Identities = 25/53 (47%), Positives = 33/53 (62%), Gaps = 3/53 (5%)
Frame = +2
Query: 313 WMSVRRQHFMMSTLKKRR--EEGSKLSKRHMMRMMKCLVVLK-EYNVLSSNEF 362
WM VRR MMST+ +RR E S + RHMM+M + + EYNVL+S+ F
Sbjct: 1262 WMIVRRPFCMMSTIWRRRCVEGSSNGAVRHMMKMRMMMSHRRNEYNVLNSSAF 1420
>AV767621
Length = 560
Score = 100 bits (248), Expect = 5e-22
Identities = 61/139 (43%), Positives = 78/139 (55%), Gaps = 23/139 (16%)
Frame = +1
Query: 3 GRAPKKSDNTKYYEILGVSKNASPDDLKKAYKKAAIKNHPDKGGDPEK-----FKELAQA 57
G +++ YY+IL V + AS DDLKKAY+K A+K HPDK + +K FK++++A
Sbjct: 31 GEGEEEAMGVDYYKILQVDRGASDDDLKKAYRKLAMKWHPDKNPNNKKEAEAKFKQISEA 210
Query: 58 YEVLSDPEKREVYDQYGEDALKEGM---------GGGGGGHDPF--------DIFSSFFG 100
Y+VLSDP+KR VYDQYGE+ LK + GG GG F DIFS FFG
Sbjct: 211 YDVLSDPQKRAVYDQYGEEGLKGQVPPPGAGGFPGGSNGGSASFRFNPRSADDIFSEFFG 390
Query: 101 -GGGFPGGGSSRGRRQRRG 118
F G G GR G
Sbjct: 391 FSSPFGGMGDMGGRAGASG 447
>AV411199
Length = 423
Score = 91.3 bits (225), Expect = 2e-19
Identities = 49/97 (50%), Positives = 64/97 (65%), Gaps = 5/97 (5%)
Frame = +2
Query: 14 YYEILGVSKNASPDDLKKAYKKAAIKNHPDKGGDPE-----KFKELAQAYEVLSDPEKRE 68
YYEIL V +A+ ++LKKAY++ A+K HPDK D + KFK ++++YEVLSDP+KR
Sbjct: 71 YYEILEVDHHATDEELKKAYRRLAMKWHPDKNPDNKNDAETKFKLISESYEVLSDPQKRA 250
Query: 69 VYDQYGEDALKEGMGGGGGGHDPFDIFSSFFGGGGFP 105
+YD+YGED LK GM G D SFF GG P
Sbjct: 251 IYDRYGEDRLKGGMPSPDAGMD------SFFRTGGGP 343
>BP059356
Length = 436
Score = 55.8 bits (133), Expect(2) = 3e-15
Identities = 33/79 (41%), Positives = 43/79 (53%), Gaps = 17/79 (21%)
Frame = +1
Query: 39 KNHPDKGGDPEKFKELAQAYEVLSDPEKREVYDQYGEDALKEGM---------GGGGGGH 89
KN +K FK++++AY+VLSDP+KR VYDQ+GE+ L + GGG GG
Sbjct: 187 KNPNNKKEAEANFKKISEAYDVLSDPQKRAVYDQFGEEGLNGQVPPPGAGGFSGGGDGGP 366
Query: 90 DPF--------DIFSSFFG 100
F DI S FFG
Sbjct: 367 ASFRFNPRSADDILSEFFG 423
Score = 42.0 bits (97), Expect(2) = 3e-15
Identities = 18/36 (50%), Positives = 26/36 (72%)
Frame = +3
Query: 7 KKSDNTKYYEILGVSKNASPDDLKKAYKKAAIKNHP 42
+K+ YY++L V ++A DDLKKAY+K A+K HP
Sbjct: 75 EKAMGVDYYKVLQVDRSAKDDDLKKAYRKLAMKWHP 182
>TC16999 similar to UP|Q9T024 (Q9T024) DNAJ-like protein
(AT4g39150/T22F8_50), partial (35%)
Length = 648
Score = 71.6 bits (174), Expect = 2e-13
Identities = 42/91 (46%), Positives = 55/91 (60%), Gaps = 4/91 (4%)
Frame = +3
Query: 14 YYEILGVSKNASPDDLKKAYKKAAIKNHPDKG-GDP---EKFKELAQAYEVLSDPEKREV 69
YY++LGV+ +AS D+KKAY A HPDK GDP E F+ L +AY+VLSDPEKRE
Sbjct: 129 YYDVLGVNVDASAADIKKAYYIKARIVHPDKNPGDPKAAENFQMLGEAYQVLSDPEKREA 308
Query: 70 YDQYGEDALKEGMGGGGGGHDPFDIFSSFFG 100
YD+ G+ + + DP +F FG
Sbjct: 309 YDKNGKAGIPQDT-----MLDPTAVFGMLFG 386
>TC16983 similar to UP|Q9ZSY2 (Q9ZSY2) ALTERED response to gravity (ARG1
protein), partial (42%)
Length = 716
Score = 68.6 bits (166), Expect = 2e-12
Identities = 34/68 (50%), Positives = 48/68 (70%), Gaps = 4/68 (5%)
Frame = +2
Query: 15 YEILGVSKNASPDDLKKAYKKAAIKNHPDKG-GDPEK---FKELAQAYEVLSDPEKREVY 70
YE+L VS++++ ++K AY+K A+K HPDK +PE FKE+A +Y +LSDPEKR Y
Sbjct: 245 YEVLSVSRDSTDQEIKTAYRKLALKYHPDKNVNNPEASELFKEVAYSYSILSDPEKRRQY 424
Query: 71 DQYGEDAL 78
D G +AL
Sbjct: 425 DSAGFEAL 448
>TC9507 similar to UP|Q8VZB6 (Q8VZB6) DnaJ-like protein, partial (40%)
Length = 905
Score = 67.0 bits (162), Expect = 5e-12
Identities = 41/117 (35%), Positives = 62/117 (52%), Gaps = 18/117 (15%)
Frame = +1
Query: 14 YYEILGVSKNASPDDLKKAYKKAAIKNHPDKG---GDPEKFKELAQAYEVLSDPEKREVY 70
YYEILG+ K+ + +D++K+Y+K ++K HPDK G E FK +++A++ L + E + Y
Sbjct: 520 YYEILGLEKSCTVEDVRKSYRKLSLKVHPDKNKAPGAEEAFKAVSKAFQCLGNEESKRKY 699
Query: 71 DQYGED--------ALKEGMGGG-------GGGHDPFDIFSSFFGGGGFPGGGSSRG 112
D GED A + G G D +IF +FF GG P +S G
Sbjct: 700 DLSGEDESVYERRAAARPGPGAARAYNGYYEADFDADEIFRNFFFGGMRPTATASFG 870
>TC10179 similar to UP|Q9SWB5 (Q9SWB5) Seed maturation protein PM37, partial
(19%)
Length = 518
Score = 65.1 bits (157), Expect = 2e-11
Identities = 34/51 (66%), Positives = 40/51 (77%), Gaps = 1/51 (1%)
Frame = +1
Query: 313 WMSVRRQHFMMSTLKKRREEGSKLSKRHMMRMMKC-LVVLKEYNVLSSNEF 362
W SV+R HFMMST KKR+ SKLS+R MMRMM C +VVLKEY+ SS+EF
Sbjct: 103 WTSVKRPHFMMST*KKRQGGDSKLSRRRMMRMMTCTVVVLKEYSAPSSDEF 255
>TC13524 similar to UP|O48678 (O48678) F3I6.4 protein, partial (19%)
Length = 485
Score = 64.3 bits (155), Expect = 3e-11
Identities = 29/61 (47%), Positives = 43/61 (69%), Gaps = 4/61 (6%)
Frame = +1
Query: 15 YEILGVSKNASPDDLKKAYKKAAIKNHPDKGGDPEK----FKELAQAYEVLSDPEKREVY 70
YE+LGVS++++ ++K AY+K A+K HPDK + K FKE+ +Y +LSDP+KR Y
Sbjct: 298 YEVLGVSRSSTDQEIKTAYRKMALKYHPDKNANDPKAADMFKEVTFSYNILSDPDKRRQY 477
Query: 71 D 71
D
Sbjct: 478 D 480
>AV775648
Length = 371
Score = 63.2 bits (152), Expect = 7e-11
Identities = 30/65 (46%), Positives = 43/65 (66%), Gaps = 3/65 (4%)
Frame = +1
Query: 10 DNTKYYEILGVSKNASPDDLKKAYKKAAIKNHPDKGGDPEKFK---ELAQAYEVLSDPEK 66
D Y+++GVS+NA+ ++KKAY K ++K+HPD DPE K ++A AYE+L D
Sbjct: 130 DEDDCYDLVGVSQNANSSEIKKAYYKLSLKHHPDTNPDPESKKLLVKVANAYEILKDEAT 309
Query: 67 REVYD 71
RE YD
Sbjct: 310 REQYD 324
>BP039305
Length = 555
Score = 57.0 bits (136), Expect = 5e-09
Identities = 31/73 (42%), Positives = 46/73 (62%), Gaps = 8/73 (10%)
Frame = +3
Query: 3 GRAPKKSD-----NTKYYEILGVSKNASPDDLKKAYKKAAIKNHPD---KGGDPEKFKEL 54
GR ++S + YY LGV K+A+ ++K AY++ A + HPD + G +KFKE+
Sbjct: 336 GRTTRRSHTVFAASADYYNTLGVPKSATVKEIKAAYRRLARQYHPDVYKEPGATDKFKEI 515
Query: 55 AQAYEVLSDPEKR 67
+ AYEVLSD +KR
Sbjct: 516 SNAYEVLSDDKKR 554
>BG662268
Length = 298
Score = 55.8 bits (133), Expect = 1e-08
Identities = 34/106 (32%), Positives = 50/106 (47%)
Frame = +1
Query: 50 KFKELAQAYEVLSDPEKREVYDQYGEDALKEGMGGGGGGHDPFDIFSSFFGGGGFPGGGS 109
KF+E++ AYEVL D KR+ YDQ G DA GG G+ D F
Sbjct: 16 KFQEVSIAYEVLKDEVKRQEYDQVGHDAYINPQAGGFEGNPFEDFVKGMF---------- 165
Query: 110 SRGRRQRRGEDVVHPLKVSLEDLYLGTSKKLSLSRNVLCSKCNGKG 155
++ G+DV +++S + G +K L+ +V C+ C G G
Sbjct: 166 ---NQKFGGDDVKTSIELSFMEAVQGCAKTLTYQTDVRCNTCGGSG 294
>CN825120
Length = 667
Score = 54.7 bits (130), Expect = 2e-08
Identities = 28/67 (41%), Positives = 38/67 (55%), Gaps = 6/67 (8%)
Frame = +3
Query: 12 TKYYEILGVSKNASPDDLKKAYKKAAIKNHPD------KGGDPEKFKELAQAYEVLSDPE 65
T YEILG++ AS ++K AY++ A HPD K +F ++ AY LSDPE
Sbjct: 201 TSLYEILGIAAAASDQEIKAAYRRLARVRHPDVAAVDRKDSSTNEFMKIHAAYSTLSDPE 380
Query: 66 KREVYDQ 72
KR YD+
Sbjct: 381 KRASYDR 401
>TC14850 homologue to UP|Q9SWB5 (Q9SWB5) Seed maturation protein PM37,
partial (25%)
Length = 577
Score = 54.3 bits (129), Expect = 3e-08
Identities = 30/49 (61%), Positives = 36/49 (73%), Gaps = 1/49 (2%)
Frame = +2
Query: 315 SVRRQHFMMSTLKKR-REEGSKLSKRHMMRMMKCLVVLKEYNVLSSNEF 362
SVRR H MMS K+R + SKLS+RHMMRMM CLVV + YN SS++F
Sbjct: 176 SVRRLHSMMSIWKRRIGGKSSKLSRRHMMRMMICLVVHRGYNAPSSDDF 322
>TC12876
Length = 820
Score = 54.3 bits (129), Expect = 3e-08
Identities = 30/82 (36%), Positives = 47/82 (56%), Gaps = 11/82 (13%)
Frame = -1
Query: 1 MFGRAPKKSDNTK-YYEILGVSKNASPDDLKKAYKKAAIKNHPDK---GGD-------PE 49
+ GR D + +Y +LG++K + +L+ AYKK A+K HPD+ G+ +
Sbjct: 274 ILGRMANGGDKSNDFYAVLGLNKECTESELRNAYKKLALKWHPDRCSASGNLKFVEEAKK 95
Query: 50 KFKELAQAYEVLSDPEKREVYD 71
KF+ + +AY VLSD KR +YD
Sbjct: 94 KFQSIQEAYSVLSDANKRLMYD 29
>TC14831 homologue to UP|P93499 (P93499) DnaJ-like protein (Fragment),
partial (63%)
Length = 851
Score = 53.1 bits (126), Expect = 7e-08
Identities = 27/64 (42%), Positives = 36/64 (56%), Gaps = 6/64 (9%)
Frame = +3
Query: 15 YEILGVSKNASPDDLKKAYKKAAIKNHPD------KGGDPEKFKELAQAYEVLSDPEKRE 68
YEILG+ AS ++K AY++ A HPD K + F ++ AY LSDPEKR
Sbjct: 249 YEILGIPAGASSQEIKAAYRRLARVCHPDVAAIDRKNSSADDFMKIHAAYSTLSDPEKRA 428
Query: 69 VYDQ 72
YD+
Sbjct: 429 TYDR 440
>BP042279
Length = 252
Score = 52.4 bits (124), Expect = 1e-07
Identities = 29/84 (34%), Positives = 41/84 (48%), Gaps = 1/84 (1%)
Frame = +1
Query: 148 CSKCNGKGSKSGA-SMTCAGCQGSGMKISMRHLGANMIQQMQHPCNECKGTGETISDKDR 206
C CNG G+K G S C+ C G G +S + QQ C+ C GTGET +
Sbjct: 13 CGTCNGTGAKPGTKSSRCSTCGGQGRVVSSTRTPLGIFQQSM-TCSSCNGTGETSTP--- 180
Query: 207 CPQCKGEKVVQQKKVLEVHVEKGM 230
C C G+ V++ K + + V G+
Sbjct: 181 CSTCSGDGRVRRTKRISLKVPAGV 252
Score = 27.7 bits (60), Expect = 3.2
Identities = 10/23 (43%), Positives = 14/23 (60%)
Frame = +1
Query: 144 RNVLCSKCNGKGSKSGASMTCAG 166
+++ CS CNG G S TC+G
Sbjct: 130 QSMTCSSCNGTGETSTPCSTCSG 198
>TC9416 similar to UP|Q9FEW7 (Q9FEW7) DnaJ like protein, partial (26%)
Length = 558
Score = 50.8 bits (120), Expect = 4e-07
Identities = 30/57 (52%), Positives = 33/57 (57%), Gaps = 2/57 (3%)
Frame = +2
Query: 252 DIVFVLQQKEHPKFKRKGEDLFVEHTLSLT--EALCGFQFALTHLDSRQLLIKSNPG 306
DIVF+ +K H F R G DL V +SLT EAL GF F LT LD R L I N G
Sbjct: 2 DIVFIFDEKPHNVFTRDGNDLVVTQKISLTEDEALTGFTFPLTTLDGRCLPIAINNG 172
Database: LJGI
Posted date: Jul 30, 2004 11:16 AM
Number of letters in database: 14,692,800
Number of sequences in database: 28,460
Lambda K H
0.316 0.136 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 6,285,888
Number of Sequences: 28460
Number of extensions: 87357
Number of successful extensions: 1164
Number of sequences better than 10.0: 116
Number of HSP's better than 10.0 without gapping: 919
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1052
length of query: 375
length of database: 4,897,600
effective HSP length: 92
effective length of query: 283
effective length of database: 2,279,280
effective search space: 645036240
effective search space used: 645036240
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)
S2: 56 (26.2 bits)
Medicago: description of AC133862.8


