

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC133709.13 - phase: 0
(557 letters)
Database: LJGI
28,460 sequences; 14,692,800 total letters
Searching..................................................done
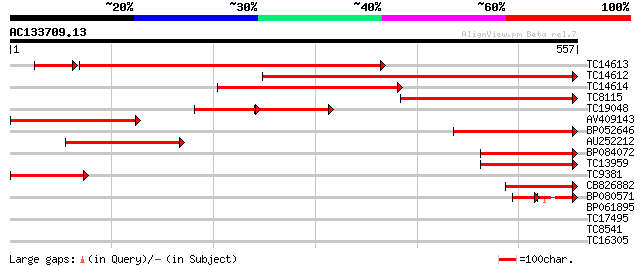
Score E
Sequences producing significant alignments: (bits) Value
TC14613 homologue to UP|PAL2_PEA (Q04593) Phenylalanine ammonia-... 522 e-164
TC14612 similar to UP|PAL1_SOYBN (P27991) Phenylalanine ammonia-... 495 e-141
TC14614 homologue to UP|PAL1_SOYBN (P27991) Phenylalanine ammoni... 320 4e-88
TC8115 similar to PIR|A24727|A24727 phenylalanine ammonia-lyase ... 271 3e-73
TC19048 homologue to PIR|A24727|A24727 phenylalanine ammonia-lya... 129 2e-59
AV409143 218 2e-57
BP052646 192 1e-49
AU252212 184 3e-47
BP084072 156 7e-39
TC13959 similar to UP|PALY_MEDSA (P27990) Phenylalanine ammonia-... 156 9e-39
TC9381 similar to UP|PALY_STYHU (P45732) Phenylalanine ammonia-l... 133 9e-32
CB826882 122 1e-28
BP080571 45 3e-12
BP061895 28 0.18
TC17495 membrane transporter [Lotus japonicus] 30 1.0
TC8541 similar to GB|AAM16209.1|20334906|AY094053 AT5g02020/T7H2... 29 1.8
TC16305 similar to UP|PAL1_SOYBN (P27991) Phenylalanine ammonia-... 29 2.3
>TC14613 homologue to UP|PAL2_PEA (Q04593) Phenylalanine ammonia-lyase 2 ,
partial (47%)
Length = 1048
Score = 522 bits (1345), Expect(2) = e-164
Identities = 258/301 (85%), Positives = 287/301 (94%)
Frame = +3
Query: 69 NGQVLNAQEAFQLAGIETGFFELQPKEGLALVNGTAVGSGLASLVLFDTNLLVVLSEILS 128
+G+VLNA+EAFQ AGI++GFFELQPKEGLALVNGTAVGSGLAS+VLFD N+L +LSE+LS
Sbjct: 144 SGKVLNAKEAFQSAGIDSGFFELQPKEGLALVNGTAVGSGLASIVLFDANILAILSEVLS 323
Query: 129 AIFAEVMLGKPQFTDHLIHKVKLHPGQIEAAAIMEHILDGSYYGKAAQKVHEINPLQKPK 188
AIFAEVM GKP+FTDHL HK+K HPGQIEAAAIMEHILDGS Y KAA+K+HE++PLQKPK
Sbjct: 324 AIFAEVMQGKPEFTDHLTHKLKHHPGQIEAAAIMEHILDGSSYMKAAKKLHEVDPLQKPK 503
Query: 189 QDRYAIRTSPQWLGPQIEVIRYATKMIEREINSVNDNPLIDVSRDKALHGGNFQGTPIGV 248
QDRYA+RTSPQWLGP IEVIR++TK IER+INSVNDNPLIDVSR+KALHGGNFQGTPIGV
Sbjct: 504 QDRYALRTSPQWLGPLIEVIRFSTKSIERDINSVNDNPLIDVSRNKALHGGNFQGTPIGV 683
Query: 249 SMDNTRLAIAAIGKLMFAQFTELVNDIYNNGLPSSLTASRNPSLDYGFKGAEVAMASYCS 308
SMDNTRLA+AAIGKLMFAQFTELV+D YNNGLPS+LTASRNPSLDYG KGAE+AMASYCS
Sbjct: 684 SMDNTRLALAAIGKLMFAQFTELVDDHYNNGLPSNLTASRNPSLDYGLKGAEIAMASYCS 863
Query: 309 ELQYLASPVTTHVQSAEQHNQDVNSLGLISARKTAEAVEIWKLMSSTFLVALCQAIDLRH 368
ELQYLA+PVTTHVQSAEQHNQDVNSLGLIS+RKT EA+EI KLMSSTFL+ALCQAIDLRH
Sbjct: 864 ELQYLANPVTTHVQSAEQHNQDVNSLGLISSRKTYEAIEILKLMSSTFLIALCQAIDLRH 1043
Query: 369 I 369
+
Sbjct: 1044L 1046
Score = 73.2 bits (178), Expect(2) = e-164
Identities = 32/42 (76%), Positives = 38/42 (90%)
Frame = +1
Query: 25 KFLNHNITPCLPLRGTITASGDLIPLSYVAGLLIGRPNSKSI 66
K +N+NITPCLPLRGT+TASGDL+PLSY+AGLL GR NS S+
Sbjct: 4 KLINNNITPCLPLRGTVTASGDLVPLSYIAGLLTGRQNSXSL 129
>TC14612 similar to UP|PAL1_SOYBN (P27991) Phenylalanine ammonia-lyase 1 ,
partial (43%)
Length = 1049
Score = 495 bits (1275), Expect = e-141
Identities = 242/309 (78%), Positives = 272/309 (87%)
Frame = +3
Query: 249 SMDNTRLAIAAIGKLMFAQFTELVNDIYNNGLPSSLTASRNPSLDYGFKGAEVAMASYCS 308
SMDNTRLA+AAIGKLMFAQFTELV+D YNNGLPS+LTASRNPSL YGFKGAE+AMASYCS
Sbjct: 3 SMDNTRLALAAIGKLMFAQFTELVDDHYNNGLPSNLTASRNPSLYYGFKGAEIAMASYCS 182
Query: 309 ELQYLASPVTTHVQSAEQHNQDVNSLGLISARKTAEAVEIWKLMSSTFLVALCQAIDLRH 368
ELQYLA+PVTTHVQSAEQHNQDVNSLGLIS+RKT EA+EI KLMSSTFL+ALCQAIDL+H
Sbjct: 183 ELQYLANPVTTHVQSAEQHNQDVNSLGLISSRKTNEAIEILKLMSSTFLIALCQAIDLKH 362
Query: 369 IEENFKSVVKNTVSQVAKRILTVGVNGELHPSRFCEKDLLNVVEGEYVFTYIDDPCSPIY 428
+EEN K V++TVSQVA+R LT GVNGELHPSRFCEKDLL VV+ E +F YIDDPC Y
Sbjct: 363 LEENLKYSVQSTVSQVAERTLTTGVNGELHPSRFCEKDLLKVVDRETLFAYIDDPCLATY 542
Query: 429 PLMQKLRYVLVDHALQNGDKEANSSTSIFQKIGAFEEELKALLPKEVENARVEIENGNPA 488
PLMQKLR VLV HAL NG+ E +S TSIFQKI FE+ELK+LLPKEVE+AR E+GNPA
Sbjct: 543 PLMQKLRQVLVXHALVNGENEKDSKTSIFQKIATFEDELKSLLPKEVESARAAYESGNPA 722
Query: 489 VPNMIKECRSYPLYKFMRETLGTSLLTGEKIRSPGEDCDKVFTAMCDGRFIDPMLDCLKE 548
+PN I ECRSYPLYKF+RE LGT LLTGEK RSPGE+ DK+FTA+C G+ IDP+++CL E
Sbjct: 723 IPNKINECRSYPLYKFVREELGTELLTGEKTRSPGEEFDKLFTAICQGKIIDPLMECLGE 902
Query: 549 WNGVPLPIC 557
WNG PLPIC
Sbjct: 903 WNGAPLPIC 929
>TC14614 homologue to UP|PAL1_SOYBN (P27991) Phenylalanine ammonia-lyase 1
, partial (26%)
Length = 550
Score = 320 bits (820), Expect = 4e-88
Identities = 162/182 (89%), Positives = 174/182 (95%)
Frame = +2
Query: 205 IEVIRYATKMIEREINSVNDNPLIDVSRDKALHGGNFQGTPIGVSMDNTRLAIAAIGKLM 264
IEVIR++TK IEREINSVNDNPLIDVSR+KALHGGNFQGTPIGVSMDNTRLA+AAIGKLM
Sbjct: 5 IEVIRFSTKSIEREINSVNDNPLIDVSRNKALHGGNFQGTPIGVSMDNTRLALAAIGKLM 184
Query: 265 FAQFTELVNDIYNNGLPSSLTASRNPSLDYGFKGAEVAMASYCSELQYLASPVTTHVQSA 324
FAQFTELV+D YNNGLPS+LTASRNPSLDYG KGAE+AMASYCSELQYLA+PVTTHVQSA
Sbjct: 185 FAQFTELVDDHYNNGLPSNLTASRNPSLDYGLKGAEIAMASYCSELQYLANPVTTHVQSA 364
Query: 325 EQHNQDVNSLGLISARKTAEAVEIWKLMSSTFLVALCQAIDLRHIEENFKSVVKNTVSQV 384
EQHNQDVNSLGLIS+RKT EAVEI KLMSSTFL+ALCQAIDLRH+EEN K VK+TVSQV
Sbjct: 365 EQHNQDVNSLGLISSRKTNEAVEILKLMSSTFLIALCQAIDLRHLEENLKYSVKSTVSQV 544
Query: 385 AK 386
AK
Sbjct: 545 AK 550
>TC8115 similar to PIR|A24727|A24727 phenylalanine ammonia-lyase - kidney
bean (fragment) {Phaseolus vulgaris;} ,
partial (34%)
Length = 760
Score = 271 bits (692), Expect = 3e-73
Identities = 128/173 (73%), Positives = 144/173 (82%)
Frame = +2
Query: 385 AKRILTVGVNGELHPSRFCEKDLLNVVEGEYVFTYIDDPCSPIYPLMQKLRYVLVDHALQ 444
A+ LT GVNGELHPSRFCEKDLL VV+ E VF YIDDPCS YPLMQKLR LVDHAL+
Sbjct: 8 ARGTLTTGVNGELHPSRFCEKDLLTVVDREDVFAYIDDPCSGTYPLMQKLRQSLVDHALE 187
Query: 445 NGDKEANSSTSIFQKIGAFEEELKALLPKEVENARVEIENGNPAVPNMIKECRSYPLYKF 504
N D E N +TSIFQKI FE+ELKA+LPKEVE+ARV +NG A+PN IKECRSYPLYKF
Sbjct: 188 NADAEKNVNTSIFQKIAIFEDELKAILPKEVESARVAYDNGESAIPNKIKECRSYPLYKF 367
Query: 505 MRETLGTSLLTGEKIRSPGEDCDKVFTAMCDGRFIDPMLDCLKEWNGVPLPIC 557
+RE LGT LLTGEK++SPGED DK+FTAMC G+ IDP+L+CL EWNG PLPIC
Sbjct: 368 VREELGTGLLTGEKVKSPGEDFDKLFTAMCQGKIIDPILECLGEWNGAPLPIC 526
>TC19048 homologue to PIR|A24727|A24727 phenylalanine ammonia-lyase -
kidney bean (fragment) {Phaseolus
vulgaris;} , partial (27%)
Length = 414
Score = 129 bits (325), Expect(2) = 2e-59
Identities = 61/78 (78%), Positives = 71/78 (90%)
Frame = +1
Query: 241 FQGTPIGVSMDNTRLAIAAIGKLMFAQFTELVNDIYNNGLPSSLTASRNPSLDYGFKGAE 300
F+ P+ V+MDNTRLA+A+IGKL+FAQF+ELVND YNNG PS+L SRNPSLDYGFKGAE
Sbjct: 181 FKEPPLRVAMDNTRLALASIGKLLFAQFSELVNDFYNNGXPSNLYGSRNPSLDYGFKGAE 360
Query: 301 VAMASYCSELQYLASPVT 318
+AMASYCSELQYLA+PVT
Sbjct: 361 IAMASYCSELQYLANPVT 414
Score = 117 bits (292), Expect(2) = 2e-59
Identities = 54/65 (83%), Positives = 61/65 (93%)
Frame = +3
Query: 182 NPLQKPKQDRYAIRTSPQWLGPQIEVIRYATKMIEREINSVNDNPLIDVSRDKALHGGNF 241
+PLQKPKQDRYA+RTS QWLGP IEV R++TK IERE+NSVNDNPLIDV+R+KALHGGNF
Sbjct: 3 DPLQKPKQDRYALRTSRQWLGPLIEVSRFSTKSIERELNSVNDNPLIDVARNKALHGGNF 182
Query: 242 QGTPI 246
QGTPI
Sbjct: 183 QGTPI 197
>AV409143
Length = 415
Score = 218 bits (555), Expect = 2e-57
Identities = 105/128 (82%), Positives = 122/128 (95%)
Frame = +1
Query: 1 MLVRINTLLQGYSGIRFEIMEAIAKFLNHNITPCLPLRGTITASGDLIPLSYVAGLLIGR 60
MLVRINTLLQGYSGIRFEI+EAI K LN+NITPCLPLRGT+TASGDL+PLSY+AGLL GR
Sbjct: 31 MLVRINTLLQGYSGIRFEILEAITKLLNNNITPCLPLRGTVTASGDLVPLSYIAGLLTGR 210
Query: 61 PNSKSIGPNGQVLNAQEAFQLAGIETGFFELQPKEGLALVNGTAVGSGLASLVLFDTNLL 120
PNSK++GP+G+VLNA+EAF+LA I++GFFELQPKEGLALVNGTAVGSGLAS+VLF+ N+L
Sbjct: 211 PNSKAVGPSGEVLNAKEAFELASIDSGFFELQPKEGLALVNGTAVGSGLASIVLFEANIL 390
Query: 121 VVLSEILS 128
VLSE+LS
Sbjct: 391 AVLSEVLS 414
>BP052646
Length = 514
Score = 192 bits (488), Expect = 1e-49
Identities = 88/121 (72%), Positives = 103/121 (84%)
Frame = -1
Query: 437 VLVDHALQNGDKEANSSTSIFQKIGAFEEELKALLPKEVENARVEIENGNPAVPNMIKEC 496
VLVDHAL NG+ E +S TSIFQKI FE+ELK+LLPKEVE+AR E+GNPA+PN I EC
Sbjct: 514 VLVDHALVNGEYEKDSKTSIFQKIATFEDELKSLLPKEVESARAAYESGNPAMPNKINEC 335
Query: 497 RSYPLYKFMRETLGTSLLTGEKIRSPGEDCDKVFTAMCDGRFIDPMLDCLKEWNGVPLPI 556
RSYPLYKF+R+ LGT LLTGEK RSPGE+CDK+FTA+C G+ IDP+L+CL EWNG PLPI
Sbjct: 334 RSYPLYKFVRKELGTELLTGEKTRSPGEECDKLFTAICQGKIIDPLLECLGEWNGAPLPI 155
Query: 557 C 557
C
Sbjct: 154 C 152
>AU252212
Length = 350
Score = 184 bits (467), Expect = 3e-47
Identities = 89/116 (76%), Positives = 105/116 (89%)
Frame = +3
Query: 56 LLIGRPNSKSIGPNGQVLNAQEAFQLAGIETGFFELQPKEGLALVNGTAVGSGLASLVLF 115
LL GRPNSK++GP+G++L A+EAF+LA I FFEL+PKEGLALVNGTAVGSGLA++VLF
Sbjct: 3 LLTGRPNSKAVGPSGEILTAKEAFELANIGHDFFELEPKEGLALVNGTAVGSGLAAIVLF 182
Query: 116 DTNLLVVLSEILSAIFAEVMLGKPQFTDHLIHKVKLHPGQIEAAAIMEHILDGSYY 171
+ N+L VLSE++SAIFAEVMLGKP+FTDHL HK+K HPGQIEAAAIMEHILDGS Y
Sbjct: 183 EANILAVLSELISAIFAEVMLGKPEFTDHLTHKLKHHPGQIEAAAIMEHILDGSSY 350
>BP084072
Length = 457
Score = 156 bits (395), Expect = 7e-39
Identities = 69/95 (72%), Positives = 81/95 (84%)
Frame = -1
Query: 463 FEEELKALLPKEVENARVEIENGNPAVPNMIKECRSYPLYKFMRETLGTSLLTGEKIRSP 522
FE+ELK+LLPKEVE+AR E+GNP + N I ECRSYPLYKF+RE LGT LLTGEK RSP
Sbjct: 457 FEDELKSLLPKEVESARAAYESGNPTISNKINECRSYPLYKFVREELGTELLTGEKSRSP 278
Query: 523 GEDCDKVFTAMCDGRFIDPMLDCLKEWNGVPLPIC 557
GE+CDK+FTA+C G+ IDP+L+CL EWNG PLPIC
Sbjct: 277 GEECDKLFTAICQGKIIDPLLECLGEWNGAPLPIC 173
>TC13959 similar to UP|PALY_MEDSA (P27990) Phenylalanine ammonia-lyase ,
partial (13%)
Length = 494
Score = 156 bits (394), Expect = 9e-39
Identities = 68/95 (71%), Positives = 81/95 (84%)
Frame = +1
Query: 463 FEEELKALLPKEVENARVEIENGNPAVPNMIKECRSYPLYKFMRETLGTSLLTGEKIRSP 522
FE+ELK+LLP EVE+AR E+GNP +PN I ECRSYPLYKF+RE LGT LLTGEK RSP
Sbjct: 4 FEDELKSLLPXEVESARAAYESGNPTIPNKINECRSYPLYKFVREELGTELLTGEKTRSP 183
Query: 523 GEDCDKVFTAMCDGRFIDPMLDCLKEWNGVPLPIC 557
GE+CD++FTA+C G+ IDP+L+CL EWNG PLPIC
Sbjct: 184 GEECDQLFTAICQGKIIDPLLECLGEWNGAPLPIC 288
>TC9381 similar to UP|PALY_STYHU (P45732) Phenylalanine ammonia-lyase ,
partial (30%)
Length = 997
Score = 133 bits (334), Expect = 9e-32
Identities = 61/77 (79%), Positives = 71/77 (91%)
Frame = +2
Query: 1 MLVRINTLLQGYSGIRFEIMEAIAKFLNHNITPCLPLRGTITASGDLIPLSYVAGLLIGR 60
MLVRINTLLQGYSGIRFEI+EAI K +N N+ PCLPLRGTITASGDL+PLSY+AGLL GR
Sbjct: 638 MLVRINTLLQGYSGIRFEILEAITKLINSNVAPCLPLRGTITASGDLVPLSYIAGLLTGR 817
Query: 61 PNSKSIGPNGQVLNAQE 77
PNSK++GP+G++L AQE
Sbjct: 818 PNSKAVGPSGEILTAQE 868
>CB826882
Length = 364
Score = 122 bits (307), Expect = 1e-28
Identities = 52/70 (74%), Positives = 61/70 (86%)
Frame = +2
Query: 488 AVPNMIKECRSYPLYKFMRETLGTSLLTGEKIRSPGEDCDKVFTAMCDGRFIDPMLDCLK 547
A+PN I ECRSYPLYKF+R+ LGT LLTGEK RSPGE+CDK+FTA+C G+ IDP+L+CL
Sbjct: 2 AMPNKINECRSYPLYKFVRKELGTELLTGEKTRSPGEECDKLFTAICQGKIIDPLLECLG 181
Query: 548 EWNGVPLPIC 557
EWNG PLPIC
Sbjct: 182 EWNGAPLPIC 211
>BP080571
Length = 338
Score = 45.4 bits (106), Expect(2) = 3e-12
Identities = 20/45 (44%), Positives = 28/45 (61%), Gaps = 6/45 (13%)
Frame = -2
Query: 519 IRSPGE------DCDKVFTAMCDGRFIDPMLDCLKEWNGVPLPIC 557
++SPG +C + + G+ IDP+L+CL EWNG PLPIC
Sbjct: 271 VKSPGHQVRSLTNCSQQYAT---GKIIDPLLECLGEWNGAPLPIC 146
Score = 43.1 bits (100), Expect(2) = 3e-12
Identities = 19/26 (73%), Positives = 22/26 (84%)
Frame = -3
Query: 495 ECRSYPLYKFMRETLGTSLLTGEKIR 520
ECRSYPLYKF+RE LGT LLT K++
Sbjct: 336 ECRSYPLYKFVREELGTELLTW*KVQ 259
Score = 36.6 bits (83), Expect = 0.011
Identities = 15/21 (71%), Positives = 18/21 (85%)
Frame = -1
Query: 516 GEKIRSPGEDCDKVFTAMCDG 536
GEK RSPGE+ DK+FTA+C G
Sbjct: 272 GEKSRSPGEEFDKLFTAICHG 210
>BP061895
Length = 246
Score = 28.5 bits (62), Expect(2) = 0.18
Identities = 10/14 (71%), Positives = 11/14 (78%)
Frame = -3
Query: 544 DCLKEWNGVPLPIC 557
+ L EWNG PLPIC
Sbjct: 196 NALGEWNGAPLPIC 155
Score = 22.7 bits (47), Expect(2) = 0.18
Identities = 8/17 (47%), Positives = 11/17 (64%)
Frame = -2
Query: 529 VFTAMCDGRFIDPMLDC 545
V T G+ IDP+L+C
Sbjct: 242 VHTQYATGKIIDPLLEC 192
>TC17495 membrane transporter [Lotus japonicus]
Length = 979
Score = 30.0 bits (66), Expect = 1.0
Identities = 24/87 (27%), Positives = 35/87 (39%), Gaps = 5/87 (5%)
Frame = +1
Query: 37 LRGTITASGDLIPLSYVAGLLIGRPNSKSIGPNGQVLNAQEAFQLAGIETGFFELQPKEG 96
+RG + L L V + +GR NS I +L + A G G +
Sbjct: 142 MRGRLWTLWILQTLGGVFCIWLGRANSLPIAVLAMILFSVGAQAACGATFGIIPFISRRS 321
Query: 97 LALVNGTA-----VGSGLASLVLFDTN 118
L +++G GSGL LV F T+
Sbjct: 322 LGIISGLTGAGGNFGSGLTQLVFFSTS 402
>TC8541 similar to GB|AAM16209.1|20334906|AY094053 AT5g02020/T7H20_70
{Arabidopsis thaliana;}, partial (44%)
Length = 869
Score = 29.3 bits (64), Expect = 1.8
Identities = 20/57 (35%), Positives = 32/57 (56%), Gaps = 2/57 (3%)
Frame = -1
Query: 471 LPKEVENARVEIENGNPAVPNMIKECRS--YPLYKFMRETLGTSLLTGEKIRSPGED 525
LP +++A + +E+G VPN++ + +PL RETL +L GE + S ED
Sbjct: 230 LPLLMKSALI*LESGG--VPNLVLDPSPPIFPLTSERRETLPVTLGDGENMDSKAED 66
>TC16305 similar to UP|PAL1_SOYBN (P27991) Phenylalanine ammonia-lyase 1 ,
partial (21%)
Length = 637
Score = 28.9 bits (63), Expect = 2.3
Identities = 13/13 (100%), Positives = 13/13 (100%)
Frame = +1
Query: 1 MLVRINTLLQGYS 13
MLVRINTLLQGYS
Sbjct: 598 MLVRINTLLQGYS 636
Database: LJGI
Posted date: Jul 30, 2004 11:16 AM
Number of letters in database: 14,692,800
Number of sequences in database: 28,460
Lambda K H
0.319 0.136 0.395
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 8,315,088
Number of Sequences: 28460
Number of extensions: 100454
Number of successful extensions: 451
Number of sequences better than 10.0: 34
Number of HSP's better than 10.0 without gapping: 450
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 451
length of query: 557
length of database: 4,897,600
effective HSP length: 95
effective length of query: 462
effective length of database: 2,193,900
effective search space: 1013581800
effective search space used: 1013581800
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 58 (26.9 bits)
Medicago: description of AC133709.13


