

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC131239.17 + phase: 0 /pseudo
(892 letters)
Database: LJGI
28,460 sequences; 14,692,800 total letters
Searching..................................................done
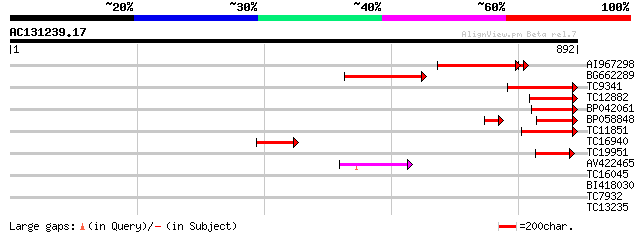
Score E
Sequences producing significant alignments: (bits) Value
AI967298 230 3e-62
BG662289 233 1e-61
TC9341 similar to UP|PNK1_ARATH (Q8L5Y9) Probable pantothenate k... 206 1e-53
TC12882 similar to UP|PNK1_ARATH (Q8L5Y9) Probable pantothenate ... 137 1e-32
BP042061 124 5e-29
BP058848 108 3e-24
TC11851 homologue to GB|AAN28872.1|23506025|AY143933 At4g35360/F... 97 9e-21
TC16940 similar to UP|PNK1_ARATH (Q8L5Y9) Probable pantothenate ... 86 3e-17
TC19951 similar to GB|AAN28872.1|23506025|AY143933 At4g35360/F23... 75 4e-14
AV422465 49 4e-06
TC16045 similar to UP|SYQ_LUPLU (P52780) Glutaminyl-tRNA synthet... 32 0.34
BI418030 28 6.4
TC7932 28 6.4
TC13235 similar to UP|Q9LQB6 (Q9LQB6) F4N2.1, partial (38%) 28 8.4
>AI967298
Length = 446
Score = 230 bits (587), Expect(2) = 3e-62
Identities = 117/131 (89%), Positives = 124/131 (94%), Gaps = 1/131 (0%)
Frame = +1
Query: 674 YHKGTIIEIYRMSRNKMRRPWRVDDFDLFKERMLGTGDKKKAPHRRALLFVDNSGADIVL 733
YH+GTIIEIYRMSRNKM+RPWRVD+FD FKERMLGTGDKK PHRRALLFVDN+GADIVL
Sbjct: 16 YHEGTIIEIYRMSRNKMQRPWRVDNFDAFKERMLGTGDKKLPPHRRALLFVDNAGADIVL 195
Query: 734 GMLPLARELLRRGTEVVLVANSLPALNDVTAMELPDIVAEAAKHCDILRRAAEAGGLLVD 793
GMLPLARELLRRGTEVVLVANSLPALNDVTAMELPDIVAEAAKHCDILRRAAE+GGLLVD
Sbjct: 196 GMLPLARELLRRGTEVVLVANSLPALNDVTAMELPDIVAEAAKHCDILRRAAESGGLLVD 375
Query: 794 AMINT-DSSKE 803
AM T DS ++
Sbjct: 376 AMTYTLDSPRQ 408
Score = 26.2 bits (56), Expect(2) = 3e-62
Identities = 12/16 (75%), Positives = 13/16 (81%)
Frame = +3
Query: 801 SKENSSSVPLMVVENG 816
S+ N SVPLMVVENG
Sbjct: 399 SQTNLPSVPLMVVENG 446
>BG662289
Length = 465
Score = 233 bits (594), Expect = 1e-61
Identities = 117/129 (90%), Positives = 122/129 (93%)
Frame = +2
Query: 528 YEPNTIDLADPSELEYWLTILSEHLPDLVDKAVASEGGTDDAKRRGDAFARAFSAHLSRL 587
YEPNTIDL+D SELEYW ILS+HLPDLVDKAVASEGGTDDAKRRGDAFARAFSAHL RL
Sbjct: 77 YEPNTIDLSDHSELEYWFKILSDHLPDLVDKAVASEGGTDDAKRRGDAFARAFSAHLVRL 256
Query: 588 MEEPSAYGKLGLANLLEMREECLREFQFVDAYRSIKQRENEASLAVLPDLFVELDSMDEE 647
MEEP+AYGKLGLANLLEMREECLREF FVDAY SIKQRENEASLAVLPDL +ELDS+DEE
Sbjct: 257 MEEPAAYGKLGLANLLEMREECLREFHFVDAYISIKQRENEASLAVLPDLLMELDSLDEE 436
Query: 648 TRLLTLIEG 656
TRLLTLIEG
Sbjct: 437 TRLLTLIEG 463
>TC9341 similar to UP|PNK1_ARATH (Q8L5Y9) Probable pantothenate kinase 1
(Pantothenic acid kinase 1) , partial (13%)
Length = 594
Score = 206 bits (524), Expect = 1e-53
Identities = 104/110 (94%), Positives = 108/110 (97%), Gaps = 1/110 (0%)
Frame = +1
Query: 784 AAEAGGLLVDAMINT-DSSKENSSSVPLMVVENGCGSPCIDLRQVSSELAAAAKDADLII 842
AAEAGGLLVDAMINT +SSKENSSSVPLMVVENGCGSPCID RQVSSELAAAAKDADLII
Sbjct: 1 AAEAGGLLVDAMINTSESSKENSSSVPLMVVENGCGSPCIDFRQVSSELAAAAKDADLII 180
Query: 843 LEGMGRSLHTNLYAQFKCDALKLAMVKNQRLAEKLIKGNIYDCICKYQPA 892
LEGMGRSLHTNLYA+FKCDALKLAMVKNQRLAEKL+KGNIYDCICKY+PA
Sbjct: 181 LEGMGRSLHTNLYARFKCDALKLAMVKNQRLAEKLVKGNIYDCICKYEPA 330
>TC12882 similar to UP|PNK1_ARATH (Q8L5Y9) Probable pantothenate kinase 1
(Pantothenic acid kinase 1) , partial (8%)
Length = 647
Score = 137 bits (344), Expect = 1e-32
Identities = 66/74 (89%), Positives = 71/74 (95%)
Frame = +2
Query: 819 SPCIDLRQVSSELAAAAKDADLIILEGMGRSLHTNLYAQFKCDALKLAMVKNQRLAEKLI 878
SPCIDL QVSSELAAAAK ADLIILEGMGRSLHTNL A+F+CDALKLAMVKNQRLAEKL+
Sbjct: 2 SPCIDLSQVSSELAAAAKGADLIILEGMGRSLHTNLNARFQCDALKLAMVKNQRLAEKLV 181
Query: 879 KGNIYDCICKYQPA 892
KGNIYDC+CKY+PA
Sbjct: 182 KGNIYDCVCKYEPA 223
>BP042061
Length = 456
Score = 124 bits (312), Expect = 5e-29
Identities = 59/72 (81%), Positives = 67/72 (92%)
Frame = -3
Query: 821 CIDLRQVSSELAAAAKDADLIILEGMGRSLHTNLYAQFKCDALKLAMVKNQRLAEKLIKG 880
CIDLR VSSELAAAAK ADLIILEGMGRSLHTNL+A+F CDA KLAMV+N RLAEKL+KG
Sbjct: 454 CIDLRPVSSELAAAAKGADLIILEGMGRSLHTNLHARFPCDAFKLAMVENPRLAEKLVKG 275
Query: 881 NIYDCICKYQPA 892
+IYDC+C+Y+PA
Sbjct: 274 DIYDCVCQYEPA 239
>BP058848
Length = 485
Score = 108 bits (271), Expect = 3e-24
Identities = 52/63 (82%), Positives = 56/63 (88%)
Frame = -2
Query: 830 ELAAAAKDADLIILEGMGRSLHTNLYAQFKCDALKLAMVKNQRLAEKLIKGNIYDCICKY 889
EL +A IILEGMGRSLHTNLYA+FKCDALKLAMVKNQRLAEKL+KGNIYDCICKY
Sbjct: 427 ELPDIVAEAAKIILEGMGRSLHTNLYARFKCDALKLAMVKNQRLAEKLVKGNIYDCICKY 248
Query: 890 QPA 892
+PA
Sbjct: 247 EPA 239
Score = 58.9 bits (141), Expect = 3e-09
Identities = 30/30 (100%), Positives = 30/30 (100%)
Frame = -2
Query: 747 TEVVLVANSLPALNDVTAMELPDIVAEAAK 776
TEVVLVANSLPALNDVTAMELPDIVAEAAK
Sbjct: 484 TEVVLVANSLPALNDVTAMELPDIVAEAAK 395
>TC11851 homologue to GB|AAN28872.1|23506025|AY143933 At4g35360/F23E12_80
{Arabidopsis thaliana;}, partial (24%)
Length = 521
Score = 97.4 bits (241), Expect = 9e-21
Identities = 48/87 (55%), Positives = 63/87 (72%)
Frame = +2
Query: 806 SSVPLMVVENGCGSPCIDLRQVSSELAAAAKDADLIILEGMGRSLHTNLYAQFKCDALKL 865
S+ L++ +G P IDL +VS ELA A DADL+ILEGMGR + TNLYAQFKCD+LK+
Sbjct: 14 STSNLLIANSGNDLPVIDLTRVSQELAYLASDADLVILEGMGRGIETNLYAQFKCDSLKI 193
Query: 866 AMVKNQRLAEKLIKGNIYDCICKYQPA 892
MVK+ ++AE + G +YDC+ KY A
Sbjct: 194 GMVKHPQVAE-FLGGRLYDCVFKYNEA 271
>TC16940 similar to UP|PNK1_ARATH (Q8L5Y9) Probable pantothenate kinase 1
(Pantothenic acid kinase 1) , partial (4%)
Length = 509
Score = 85.9 bits (211), Expect = 3e-17
Identities = 39/66 (59%), Positives = 50/66 (75%)
Frame = +2
Query: 389 VQYWSNGEAQAMFLRHEGFLGALGAFMSYEKHGLDDLMVHQLVERFPMGAPYTGGKIHGP 448
V +WS GEAQAMFLRHEGFLGA+GAFM +KHGL +L+V+Q+ ++ P + KIHGP
Sbjct: 14 VNFWSKGEAQAMFLRHEGFLGAMGAFMRSDKHGLKELLVNQVAQQTPTKFSFAVDKIHGP 193
Query: 449 PLGDLN 454
G+LN
Sbjct: 194 VDGELN 211
>TC19951 similar to GB|AAN28872.1|23506025|AY143933 At4g35360/F23E12_80
{Arabidopsis thaliana;}, partial (15%)
Length = 393
Score = 75.5 bits (184), Expect = 4e-14
Identities = 35/61 (57%), Positives = 47/61 (76%)
Frame = +1
Query: 828 SSELAAAAKDADLIILEGMGRSLHTNLYAQFKCDALKLAMVKNQRLAEKLIKGNIYDCIC 887
S E+A A DADL+ILEGMGR + TNL+A+FKCD+LK+AMVK+ +AE + +YDC+
Sbjct: 1 SEEIAFHANDADLVILEGMGRGIETNLFAEFKCDSLKIAMVKHPEVAE-FLGSRLYDCVI 177
Query: 888 K 888
K
Sbjct: 178 K 180
>AV422465
Length = 413
Score = 48.9 bits (115), Expect = 4e-06
Identities = 33/124 (26%), Positives = 62/124 (49%), Gaps = 9/124 (7%)
Frame = +3
Query: 519 FPLLADPKLYEPNTIDLADPSELEY--------WLTILSEHLPDLVDKAVASEGGTDDAK 570
FPLL + Y +TI PS+ + W+ +L +P +A S+ DA
Sbjct: 39 FPLLKNDN-YTASTIPWRFPSDDPHIPTPTELSWINLLLNTIPSFKKRA-ESDTSVPDAA 212
Query: 571 RRGDAFARAFSAHLSRLMEEPSAYG-KLGLANLLEMREECLREFQFVDAYRSIKQRENEA 629
+ + FA+ ++ L ++P++ G + L +RE+ LRE F+D ++ +K+ EN
Sbjct: 213 NKAEIFAQRYAKILEDFKKDPASQGGPPDVLFLCRLREQELRELGFIDIFKKVKEEENAK 392
Query: 630 SLAV 633
++A+
Sbjct: 393 AIAL 404
>TC16045 similar to UP|SYQ_LUPLU (P52780) Glutaminyl-tRNA synthetase
(Glutamine--tRNA ligase) (GlnRS) , partial (14%)
Length = 607
Score = 32.3 bits (72), Expect = 0.34
Identities = 24/71 (33%), Positives = 37/71 (51%), Gaps = 2/71 (2%)
Frame = +3
Query: 508 GVLHLV--PTLEVFPLLADPKLYEPNTIDLADPSELEYWLTILSEHLPDLVDKAVASEGG 565
GVLH V P+ V PL + +L+E + +P+ELE WL L+ H ++ A G
Sbjct: 18 GVLHWVAQPSSGVEPLKVEVRLFERLFLS-ENPAELENWLGDLNPHSKVVIPNAYG--GS 188
Query: 566 TDDAKRRGDAF 576
+ + GD+F
Sbjct: 189 SLQGSKVGDSF 221
>BI418030
Length = 488
Score = 28.1 bits (61), Expect = 6.4
Identities = 18/39 (46%), Positives = 20/39 (51%), Gaps = 1/39 (2%)
Frame = -1
Query: 778 CDILRRAAEAGGLLV-DAMINTDSSKENSSSVPLMVVEN 815
C I A GGLLV + INT S VPLMV E+
Sbjct: 278 CSIFSTRATYGGLLVSEGRINTQSPALTILLVPLMVYES 162
>TC7932
Length = 392
Score = 28.1 bits (61), Expect = 6.4
Identities = 12/34 (35%), Positives = 19/34 (55%)
Frame = -2
Query: 11 QQQVLGIDESEGHNNTNNMAPTGNPIHRSSSRPQ 44
QQ +GI + N + +PT + H+SS+ PQ
Sbjct: 103 QQSCIGIPQQSQQVNQSGPSPTASTAHQSSALPQ 2
>TC13235 similar to UP|Q9LQB6 (Q9LQB6) F4N2.1, partial (38%)
Length = 467
Score = 27.7 bits (60), Expect = 8.4
Identities = 12/23 (52%), Positives = 16/23 (69%)
Frame = +2
Query: 763 TAMELPDIVAEAAKHCDILRRAA 785
TA + D AEA+KH DI++ AA
Sbjct: 182 TAKKSKDFAAEASKHADIIKTAA 250
Database: LJGI
Posted date: Jul 30, 2004 11:16 AM
Number of letters in database: 14,692,800
Number of sequences in database: 28,460
Lambda K H
0.319 0.137 0.397
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 12,791,982
Number of Sequences: 28460
Number of extensions: 154611
Number of successful extensions: 728
Number of sequences better than 10.0: 28
Number of HSP's better than 10.0 without gapping: 720
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 727
length of query: 892
length of database: 4,897,600
effective HSP length: 98
effective length of query: 794
effective length of database: 2,108,520
effective search space: 1674164880
effective search space used: 1674164880
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 59 (27.3 bits)
Medicago: description of AC131239.17


