

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC126785.11 + phase: 0
(538 letters)
Database: LJGI
28,460 sequences; 14,692,800 total letters
Searching..................................................done
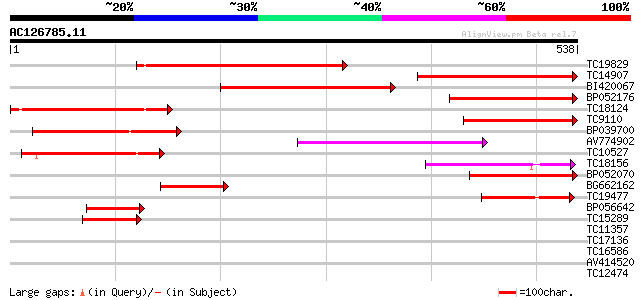
Score E
Sequences producing significant alignments: (bits) Value
TC19829 similar to GB|BAB10858.1|10177467|AB009053 permease 1 {A... 285 1e-77
TC14907 similar to GB|BAB10858.1|10177467|AB009053 permease 1 {A... 256 6e-69
BI420067 254 3e-68
BP052176 231 3e-61
TC18124 similar to GB|BAB10858.1|10177467|AB009053 permease 1 {A... 216 6e-57
TC9110 similar to GB|BAB10858.1|10177467|AB009053 permease 1 {Ar... 176 7e-45
BP039700 153 8e-38
AV774902 134 5e-32
TC10527 123 7e-29
TC18156 122 2e-28
BP052070 119 9e-28
BG662162 103 5e-23
TC19477 101 3e-22
BP056642 80 8e-16
TC15289 weakly similar to UP|AAR18373 (AAR18373) Nucleobase-asco... 52 3e-07
TC11357 37 0.011
TC17136 weakly similar to UP|AAR18373 (AAR18373) Nucleobase-asco... 34 0.052
TC16586 weakly similar to UP|Q93VQ9 (Q93VQ9) At1g31020/F17F8_6 (... 32 0.34
AV414520 31 0.44
TC12474 weakly similar to UP|O82576 (O82576) Blue copper protein... 30 1.3
>TC19829 similar to GB|BAB10858.1|10177467|AB009053 permease 1 {Arabidopsis
thaliana;} , partial (37%)
Length = 600
Score = 285 bits (729), Expect = 1e-77
Identities = 138/200 (69%), Positives = 169/200 (84%)
Frame = +3
Query: 121 LASRYDDDIMHPREKFKRIMRGTQGALIVASSLQIIVGFSGLWCHVVRFISPLSAVPLVA 180
LA R+ D+ P EKFK+IMR QGALIVAS+LQI++GFSGLW +V RF+SPLSAVPLV+
Sbjct: 3 LAGRFSDE-PDPIEKFKKIMRAIQGALIVASTLQIVLGFSGLWRNVARFLSPLSAVPLVS 179
Query: 181 LTGFGLYELGFPMLAKCIEIGLPEIVILVFLSQFMPHMMKGGRHIFARFAVIFSVIIVWV 240
L GFGLYELGFP +AKCIEIGLPE+V++VF+SQ++PH++ G+HIF RFAVIF+++IVW+
Sbjct: 180 LVGFGLYELGFPGVAKCIEIGLPELVLIVFVSQYVPHVLHKGKHIFDRFAVIFTIVIVWI 359
Query: 241 YAIILTGCGAYKNAEHETQDTCRTDRAGLIHGASWISPPIPFRWGAPTFDAGEAFAMMAA 300
YA +LT GAY NA H+TQ +CRTDRAGLI A WI P PF+WGAP+FDAGEAFAMM A
Sbjct: 360 YAHLLTVGGAYNNAAHKTQISCRTDRAGLIDNAPWIRVPYPFQWGAPSFDAGEAFAMMMA 539
Query: 301 SFVAQIESTGGFIAVARFAS 320
+FVA +ESTG FIAV R+AS
Sbjct: 540 AFVALVESTGAFIAVYRYAS 599
>TC14907 similar to GB|BAB10858.1|10177467|AB009053 permease 1 {Arabidopsis
thaliana;} , partial (28%)
Length = 734
Score = 256 bits (654), Expect = 6e-69
Identities = 115/151 (76%), Positives = 131/151 (86%)
Frame = +3
Query: 388 KFGAVFASIPMPIVAALYCLLFSQVGSAGLSFLQFCNLNSFRTKFIIGFSIFMGFSVPQY 447
KFGAVFASIP P++AALYCL F VG+ GLSFLQFCNLNSFRTKF++GFSIF+G S+PQY
Sbjct: 3 KFGAVFASIPPPMIAALYCLFFGYVGAGGLSFLQFCNLNSFRTKFVLGFSIFLGLSIPQY 182
Query: 448 FKEYTAIKQYGPVHTNARWFNDMINVPFSSGAFVAGILALFFDVTLHKSDNQTRKDRGMH 507
F EYTAI YGPVHT ARWFND++NVPF S AFVAG++A F D TLHK ++ RKDRG H
Sbjct: 183 FNEYTAINGYGPVHTGARWFNDIVNVPFQSKAFVAGVVAYFLDNTLHKKESAIRKDRGKH 362
Query: 508 WWDRFSSFKTDTRSEEFYSLPFNLNKFFPSV 538
WWD++ SFKTDTRSEEFYSLPFNLNK+FPSV
Sbjct: 363 WWDKYRSFKTDTRSEEFYSLPFNLNKYFPSV 455
>BI420067
Length = 499
Score = 254 bits (648), Expect = 3e-68
Identities = 120/166 (72%), Positives = 142/166 (85%)
Frame = +1
Query: 201 GLPEIVILVFLSQFMPHMMKGGRHIFARFAVIFSVIIVWVYAIILTGCGAYKNAEHETQD 260
GLPE++ILVF+SQ++PH + G+H+F RFAV+ ++ IVW+YA ILT GAY +A +TQ
Sbjct: 1 GLPELIILVFVSQYVPHALHSGKHVFDRFAVLITIAIVWLYAYILTVGGAYNHAARKTQT 180
Query: 261 TCRTDRAGLIHGASWISPPIPFRWGAPTFDAGEAFAMMAASFVAQIESTGGFIAVARFAS 320
TCRTDRAGLI A WI P PF+WGAP+FDAGEAFAMM ASFVA +ES+G F AV RFAS
Sbjct: 181 TCRTDRAGLIDAAPWIRVPYPFQWGAPSFDAGEAFAMMMASFVALVESSGAFTAVYRFAS 360
Query: 321 ATPVPPSVLSRGIGWQGVGILLSGIFGTGNGSSVSIENAGLLALTR 366
ATP+PPS+LSRGIGWQGVGILLSG+FGTGNGSSVS+ENAGLLALTR
Sbjct: 361 ATPLPPSILSRGIGWQGVGILLSGLFGTGNGSSVSVENAGLLALTR 498
>BP052176
Length = 529
Score = 231 bits (588), Expect = 3e-61
Identities = 106/121 (87%), Positives = 113/121 (92%)
Frame = -2
Query: 418 SFLQFCNLNSFRTKFIIGFSIFMGFSVPQYFKEYTAIKQYGPVHTNARWFNDMINVPFSS 477
SFLQFCNLNSFRTKFI+GFSIFMGFS+PQYFKEYTA+K YGPVHT+ARWFNDMINVPFSS
Sbjct: 528 SFLQFCNLNSFRTKFILGFSIFMGFSIPQYFKEYTALKHYGPVHTHARWFNDMINVPFSS 349
Query: 478 GAFVAGILALFFDVTLHKSDNQTRKDRGMHWWDRFSSFKTDTRSEEFYSLPFNLNKFFPS 537
AFVAG LALF D TL K+D+QTRKDRG HWWDRF S+KTDTRSEEFYSLPFNLNKFFPS
Sbjct: 348 EAFVAGSLALFLDATLRKNDSQTRKDRGAHWWDRFRSYKTDTRSEEFYSLPFNLNKFFPS 169
Query: 538 V 538
V
Sbjct: 168 V 166
>TC18124 similar to GB|BAB10858.1|10177467|AB009053 permease 1 {Arabidopsis
thaliana;} , partial (26%)
Length = 836
Score = 216 bits (551), Expect = 6e-57
Identities = 115/155 (74%), Positives = 128/155 (82%), Gaps = 1/155 (0%)
Frame = +1
Query: 1 MAGGGGGGGATPPPKQDEFQPHPVKDQLPNVSYCITSPPPWPEA-IMLGFQHYLVMLGTT 59
MAGGGGGG P PK DE QPHP KDQLPNVSYCITSPP LGFQHYLVMLGTT
Sbjct: 379 MAGGGGGGA--PAPKADEPQPHPAKDQLPNVSYCITSPPSMA*GHSCLGFQHYLVMLGTT 552
Query: 60 VLIPTALVSQMGGGNEEKAMLIQNHLFVAGINTLIQTLFGTRLPAVIGGSFTFVPTTISI 119
VLIPTALV QMGGGN+EKA +IQ LFVAGINT +Q++FGTRLPAV+GGS+T+VPTTISI
Sbjct: 553 VLIPTALVPQMGGGNDEKAKVIQTLLFVAGINT*LQSMFGTRLPAVMGGSYTYVPTTISI 732
Query: 120 ILASRYDDDIMHPREKFKRIMRGTQGALIVASSLQ 154
ILA R+ D+ P EKFKRI+R +QGALIVAS+LQ
Sbjct: 733 ILAGRFSDE-PDPIEKFKRIVRASQGALIVASTLQ 834
>TC9110 similar to GB|BAB10858.1|10177467|AB009053 permease 1 {Arabidopsis
thaliana;} , partial (20%)
Length = 595
Score = 176 bits (447), Expect = 7e-45
Identities = 78/108 (72%), Positives = 90/108 (83%)
Frame = +1
Query: 431 KFIIGFSIFMGFSVPQYFKEYTAIKQYGPVHTNARWFNDMINVPFSSGAFVAGILALFFD 490
KFI+GFSIF+G S+PQYF EYTAI +GPVHT ARWFND+INVPF S FV G++A F D
Sbjct: 1 KFILGFSIFLGLSIPQYFNEYTAINGFGPVHTGARWFNDIINVPFQSKPFVGGVVAYFLD 180
Query: 491 VTLHKSDNQTRKDRGMHWWDRFSSFKTDTRSEEFYSLPFNLNKFFPSV 538
TL+K D+ RKDRG HWWD++ SFK DTRSEEFYSLPFNLNK+FPSV
Sbjct: 181 NTLYKRDSAIRKDRGKHWWDKYKSFKGDTRSEEFYSLPFNLNKYFPSV 324
>BP039700
Length = 506
Score = 153 bits (386), Expect = 8e-38
Identities = 74/142 (52%), Positives = 103/142 (72%)
Frame = +2
Query: 22 HPVKDQLPNVSYCITSPPPWPEAIMLGFQHYLVMLGTTVLIPTALVSQMGGGNEEKAMLI 81
H +QL ++ YC+ S PPW E ++L FQ+Y++MLGT+V+IP+ +V MGG + +KA +I
Sbjct: 74 HLPMEQLQDLEYCLDSNPPWAETVLLAFQNYILMLGTSVMIPSLIVPAMGGSDGDKARVI 253
Query: 82 QNHLFVAGINTLIQTLFGTRLPAVIGGSFTFVPTTISIILASRYDDDIMHPREKFKRIMR 141
Q LFV+GINTL+Q LFGTRLPAV+GGSF +V ++ I+ I P E+F + MR
Sbjct: 254 QTLLFVSGINTLLQALFGTRLPAVVGGSFAYV-VPVAYIITDSSLQKITDPHERFIQTMR 430
Query: 142 GTQGALIVASSLQIIVGFSGLW 163
QGALIVASS+QII+G+S +W
Sbjct: 431 AIQGALIVASSIQIILGYSQVW 496
>AV774902
Length = 594
Score = 134 bits (336), Expect = 5e-32
Identities = 65/180 (36%), Positives = 105/180 (58%)
Frame = +2
Query: 274 SWISPPIPFRWGAPTFDAGEAFAMMAASFVAQIESTGGFIAVARFASATPVPPSVLSRGI 333
+W+ P P +WG P F + M+ S VA ++S G + ++ P P V+SRG+
Sbjct: 38 AWVRFPYP*QWGIPIFHFKTSIIMVIVSLVASVDSVGTYRTASQQVDLRPPTPRVVSRGM 217
Query: 334 GWQGVGILLSGIFGTGNGSSVSIENAGLLALTRVGSRRVVQISAGFMIFFSILGKFGAVF 393
+G + +G++G+G GS+ EN + T+V SR+ V + A F+I FS +GK GA+
Sbjct: 218 ALEGFCSIWAGLWGSGTGSTTLTENIHTINSTKVASRKAVVLGAAFLILFS*IGKVGALL 397
Query: 394 ASIPMPIVAALYCLLFSQVGSAGLSFLQFCNLNSFRTKFIIGFSIFMGFSVPQYFKEYTA 453
ASIP+ + A++ C +++ GLS LQ+ SFR I+G S+F+G S+P Y ++Y A
Sbjct: 398 ASIPLSLAASVLCFMWALTVGLGLSTLQYGQSASFRNMTIVGVSLFLGMSIPSYIQQYQA 577
>TC10527
Length = 529
Score = 123 bits (309), Expect = 7e-29
Identities = 66/139 (47%), Positives = 88/139 (62%), Gaps = 3/139 (2%)
Frame = +2
Query: 12 PPPKQDEFQPHPV---KDQLPNVSYCITSPPPWPEAIMLGFQHYLVMLGTTVLIPTALVS 68
PPP P +QL + YCI S P WPEA++LGFQHY+VMLGTTVLI T+LV+
Sbjct: 116 PPPPNFGLSRGPTWAPAEQLLQLDYCIHSNPSWPEALLLGFQHYIVMLGTTVLIATSLVN 295
Query: 69 QMGGGNEEKAMLIQNHLFVAGINTLIQTLFGTRLPAVIGGSFTFVPTTISIILASRYDDD 128
QMGG + +KA +IQ L ++ INTL+QT FG+RLP V+GGS ++ +S+I D
Sbjct: 296 QMGGTHGDKARVIQTMLIMSAINTLLQTWFGSRLPTVMGGSVAYILPVMSVI-NDYTDKT 472
Query: 129 IMHPREKFKRIMRGTQGAL 147
E+F +R QG+L
Sbjct: 473 FSSEHERFTYTIRTVQGSL 529
>TC18156
Length = 531
Score = 122 bits (305), Expect = 2e-28
Identities = 64/146 (43%), Positives = 85/146 (57%), Gaps = 3/146 (2%)
Frame = +1
Query: 395 SIPMPIVAALYCLLFSQVGSAGLSFLQFCNLNSFRTKFIIGFSIFMGFSVPQYFKEYTAI 454
SIP+ I AA+YC+LF V + G+SF+QF +S +I+G S+F+G S+P YF +
Sbjct: 1 SIPLSIFAAIYCVLFGIVAAIGISFIQFSYYDSMSNIYILGLSLFLGISIPLYFIMNISP 180
Query: 455 KQYGPVHTNARWFNDMINVPFSSGAFVAGILALFFDVTL---HKSDNQTRKDRGMHWWDR 511
GPV TN WFND++N FSS VA I+ D TL HK D DRG+ WW
Sbjct: 181 DGRGPVRTNGGWFNDILNTIFSSPPTVAIIVGTVLDNTLDVKHKED-----DRGLTWWRP 345
Query: 512 FSSFKTDTRSEEFYSLPFNLNKFFPS 537
F K D R++EFY P L ++ PS
Sbjct: 346 FQHRKGDVRNDEFYRFPLGLTEYIPS 423
>BP052070
Length = 480
Score = 119 bits (299), Expect = 9e-28
Identities = 54/102 (52%), Positives = 73/102 (70%)
Frame = -2
Query: 437 SIFMGFSVPQYFKEYTAIKQYGPVHTNARWFNDMINVPFSSGAFVAGILALFFDVTLHKS 496
S+F+G SVPQYF E + +GPVHT + FN+++ V FSS A VA I+A DVTL +
Sbjct: 479 SLFLGLSVPQYFNEQLLLSGHGPVHTGSIAFNNIVQVIFSSPATVAIIVAYLLDVTLSRG 300
Query: 497 DNQTRKDRGMHWWDRFSSFKTDTRSEEFYSLPFNLNKFFPSV 538
+ TR+D G HWW++F +F D R+EEFYSLP NLN++FPS+
Sbjct: 299 HSSTRRDSGRHWWEKFRNFDQDHRNEEFYSLPMNLNRYFPSL 174
>BG662162
Length = 204
Score = 103 bits (258), Expect = 5e-23
Identities = 47/64 (73%), Positives = 59/64 (91%)
Frame = +3
Query: 144 QGALIVASSLQIIVGFSGLWCHVVRFISPLSAVPLVALTGFGLYELGFPMLAKCIEIGLP 203
+GALIVAS+LQI++GFSGLW +V RF+SPLS VPLV+L GFGLYELGFP +AKC++IGLP
Sbjct: 12 EGALIVASTLQIVLGFSGLWRNVARFLSPLSVVPLVSLVGFGLYELGFPGVAKCVQIGLP 191
Query: 204 EIVI 207
E++I
Sbjct: 192 ELII 203
>TC19477
Length = 568
Score = 101 bits (252), Expect = 3e-22
Identities = 49/89 (55%), Positives = 60/89 (67%)
Frame = +1
Query: 448 FKEYTAIKQYGPVHTNARWFNDMINVPFSSGAFVAGILALFFDVTLHKSDNQTRKDRGMH 507
F+EYT + P HT WFND +N F S VA I+A+F D TL D+ KDRGM
Sbjct: 1 FREYTIRPLHVPAHTKXGWFNDFLNTIFYSPPTVALIVAVFLDNTLDYKDSA--KDRGMP 174
Query: 508 WWDRFSSFKTDTRSEEFYSLPFNLNKFFP 536
WW +F +FK D+R+EEFYSLPFNLN+FFP
Sbjct: 175 WWAKFRAFKADSRNEEFYSLPFNLNRFFP 261
>BP056642
Length = 495
Score = 80.1 bits (196), Expect = 8e-16
Identities = 39/55 (70%), Positives = 47/55 (84%)
Frame = +1
Query: 74 NEEKAMLIQNHLFVAGINTLIQTLFGTRLPAVIGGSFTFVPTTISIILASRYDDD 128
N EKA +I+ L VAGINTL+QTLFG+RLPAVIGGS+T+V TTISIILA R+ D+
Sbjct: 19 NNEKARVIETLLXVAGINTLLQTLFGSRLPAVIGGSYTYVATTISIILAGRFSDE 183
>TC15289 weakly similar to UP|AAR18373 (AAR18373) Nucleobase-ascorbate
transporter 11, partial (9%)
Length = 583
Score = 51.6 bits (122), Expect = 3e-07
Identities = 25/56 (44%), Positives = 35/56 (61%)
Frame = +2
Query: 70 MGGGNEEKAMLIQNHLFVAGINTLIQTLFGTRLPAVIGGSFTFVPTTISIILASRY 125
MGG + A +I LF++GI T++ + FGTRLP V GGSF ++ + II A Y
Sbjct: 2 MGGTDTNTANVIPTMLFLSGITTILHSYFGTRLPLVQGGSFVYLEPALVIINAQEY 169
>TC11357
Length = 580
Score = 36.6 bits (83), Expect = 0.011
Identities = 14/19 (73%), Positives = 17/19 (88%)
Frame = +3
Query: 518 DTRSEEFYSLPFNLNKFFP 536
D +EEFY+LPFNLN+FFP
Sbjct: 9 DDSNEEFYTLPFNLNRFFP 65
>TC17136 weakly similar to UP|AAR18373 (AAR18373) Nucleobase-ascorbate
transporter 11, partial (11%)
Length = 512
Score = 34.3 bits (77), Expect = 0.052
Identities = 23/84 (27%), Positives = 35/84 (41%)
Frame = +3
Query: 445 PQYFKEYTAIKQYGPVHTNARWFNDMINVPFSSGAFVAGILALFFDVTLHKSDNQTRKDR 504
P Y Y A GP H+ + + IN S V ++A D T+ S +++R
Sbjct: 3 PSYLIPYAAASS-GPSHSGIKPLDFAINALLSMNMVVTLLVAFLLDNTVPGS----QEER 167
Query: 505 GMHWWDRFSSFKTDTRSEEFYSLP 528
G++ W + D YSLP
Sbjct: 168 GVYTWSKAEDITADPSLPSEYSLP 239
>TC16586 weakly similar to UP|Q93VQ9 (Q93VQ9) At1g31020/F17F8_6 (Thioredoxin
o) (At2g20920/F5H14.11), partial (35%)
Length = 898
Score = 31.6 bits (70), Expect = 0.34
Identities = 15/45 (33%), Positives = 26/45 (57%)
Frame = +2
Query: 129 IMHPREKFKRIMRGTQGALIVASSLQIIVGFSGLWCHVVRFISPL 173
+++ E+F RI+ Q +SL I+ F+ +WC +FISP+
Sbjct: 290 LVNSEEEFNRILTKIQD-----NSLHAIIYFTAVWCGPCKFISPI 409
>AV414520
Length = 409
Score = 31.2 bits (69), Expect = 0.44
Identities = 14/41 (34%), Positives = 19/41 (46%)
Frame = +3
Query: 3 GGGGGGGATPPPKQDEFQPHPVKDQLPNVSYCITSPPPWPE 43
GG GGG PPP D HP+ ++ + PP P+
Sbjct: 15 GGEGGGQTLPPPTTDHTLSHPLLHHQHHLILTLQPTPPPPK 137
>TC12474 weakly similar to UP|O82576 (O82576) Blue copper protein
(Fragment), partial (44%)
Length = 818
Score = 29.6 bits (65), Expect = 1.3
Identities = 14/38 (36%), Positives = 17/38 (43%)
Frame = +1
Query: 11 TPPPKQDEFQPHPVKDQLPNVSYCITSPPPWPEAIMLG 48
TPPP P P P+ S +T PP A+ LG
Sbjct: 517 TPPPPSSTTTPSPSSPTSPSPSSTVTPPPKDSAAVSLG 630
Database: LJGI
Posted date: Jul 30, 2004 11:16 AM
Number of letters in database: 14,692,800
Number of sequences in database: 28,460
Lambda K H
0.327 0.142 0.443
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 9,813,496
Number of Sequences: 28460
Number of extensions: 141905
Number of successful extensions: 1898
Number of sequences better than 10.0: 61
Number of HSP's better than 10.0 without gapping: 1501
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1798
length of query: 538
length of database: 4,897,600
effective HSP length: 95
effective length of query: 443
effective length of database: 2,193,900
effective search space: 971897700
effective search space used: 971897700
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 15 ( 7.1 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.7 bits)
S2: 57 (26.6 bits)
Medicago: description of AC126785.11


