

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC121233.14 - phase: 0
(275 letters)
Database: LJGI
28,460 sequences; 14,692,800 total letters
Searching..................................................done
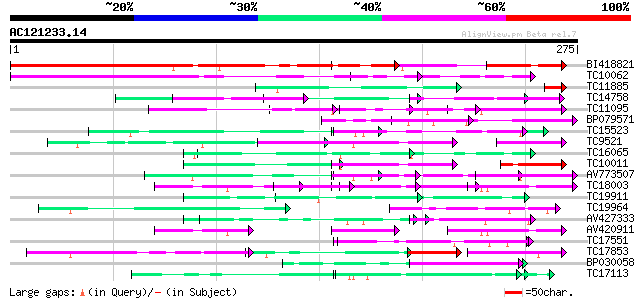
Score E
Sequences producing significant alignments: (bits) Value
BI418821 244 1e-65
TC10062 similar to BAD08700 (BAD08700) Cold shock domain protein... 142 8e-35
TC11885 similar to UP|Q8LF59 (Q8LF59) DNA-binding protein, parti... 54 1e-09
TC14758 similar to UP|O24601 (O24601) Glycine-rich RNA binding p... 59 1e-09
TC11095 similar to PIR|T05112|T05112 splicing factor 9G8-like SR... 57 3e-09
BP079571 57 4e-09
TC15523 similar to UP|GRPA_MAIZE (P10979) Glycine-rich RNA-bindi... 55 1e-08
TC9521 similar to UP|Q9FYA7 (Q9FYA7) Splicing factor RSZ33, part... 54 3e-08
TC16065 weakly similar to UP|Q9SBM1 (Q9SBM1) Hydroxyproline-rich... 52 1e-07
TC10011 similar to UP|Q9FYA7 (Q9FYA7) Splicing factor RSZ33, par... 52 1e-07
AV773507 50 5e-07
TC18003 similar to PIR|T05112|T05112 splicing factor 9G8-like SR... 46 6e-06
TC19911 weakly similar to PIR|A44805|A44805 eggshell protein pre... 46 6e-06
TC19964 similar to UP|Q40363 (Q40363) NuM1 protein, partial (11%) 46 8e-06
AV427333 45 1e-05
AV420911 45 2e-05
TC17551 weakly similar to UP|AAR24662 (AAR24662) At5g26960, part... 44 2e-05
TC17853 similar to UP|Q42412 (Q42412) RNA-binding protein RZ-1, ... 44 4e-05
BP030058 43 5e-05
TC17113 weakly similar to UP|Q9LFU8 (Q9LFU8) Proline-rich protei... 42 9e-05
>BI418821
Length = 614
Score = 244 bits (623), Expect = 1e-65
Identities = 128/196 (65%), Positives = 139/196 (70%), Gaps = 7/196 (3%)
Frame = +2
Query: 1 MAEERISGVVQWFNNTKGFGFIKPDDDSEDLFVHQSDIRSEGFRSLSEGDRVEFTIADGD 60
MAEER SGVVQWFNN KGFGFIKPDD EDLFVHQS IRS+G+R+L EGD VEF+IA GD
Sbjct: 65 MAEERFSGVVQWFNNVKGFGFIKPDDGGEDLFVHQSSIRSDGYRTLLEGDLVEFSIATGD 244
Query: 61 NGKTKAVDVTGPKGEPLQ-VRQDNHGGGGGGRGFRGGERRN------GGGGCYTCGDTGH 113
N KTKAVDVTGP G LQ R+D+ G G G+RGGERRN GGGGCY CGDTGH
Sbjct: 245 NDKTKAVDVTGPNGAALQPTRKDSAPRGFG--GWRGGERRNGGGGGGGGGGCYNCGDTGH 418
Query: 114 IARDCDRSDRNDRNDRSGGGGGGDRDRACYTCGSFEHFARDCMRGGGNNNNGGGGYGGGG 173
+ARDC RS+ N GGGGGG ACY CG H ARDC R NNN+GGGG G
Sbjct: 419 LARDCHRSNNN------GGGGGG---AACYNCGDAGHLARDCNR--SNNNSGGGGAG--- 556
Query: 174 TSCYRCGGVGHIARDC 189
CY CG GH+ARDC
Sbjct: 557 --CYNCGDTGHLARDC 598
Score = 107 bits (266), Expect = 3e-24
Identities = 53/116 (45%), Positives = 60/116 (51%), Gaps = 2/116 (1%)
Frame = +2
Query: 157 RGGGNNNNGGGGYGGGGTSCYRCGGVGHIARDC--ATPSSGGGGGGACYKCGEVGHIARD 214
RGG N GGGG GGGG CY CG GH+ARDC + + GGGGG ACY CG+ GH+ARD
Sbjct: 341 RGGERRNGGGGGGGGGG--CYNCGDTGHLARDCHRSNNNGGGGGGAACYNCGDAGHLARD 514
Query: 215 CSNEGGRFDGGNGRYDDGNGRFGGGNRRFGSGGGGHDGGKGTCFNCGKAGHFARDC 270
C+ N G GG G C+NCG GH ARDC
Sbjct: 515 CNR---------------------SNNNSGGGGAG-------CYNCGDTGHLARDC 598
Score = 50.8 bits (120), Expect = 2e-07
Identities = 23/39 (58%), Positives = 25/39 (63%)
Frame = +2
Query: 232 GNGRFGGGNRRFGSGGGGHDGGKGTCFNCGKAGHFARDC 270
G G + GG RR G GGGG GG C+NCG GH ARDC
Sbjct: 326 GFGGWRGGERRNGGGGGGGGGG---CYNCGDTGHLARDC 433
>TC10062 similar to BAD08700 (BAD08700) Cold shock domain protein 2, partial
(66%)
Length = 481
Score = 142 bits (357), Expect = 8e-35
Identities = 85/200 (42%), Positives = 103/200 (51%)
Frame = +3
Query: 1 MAEERISGVVQWFNNTKGFGFIKPDDDSEDLFVHQSDIRSEGFRSLSEGDRVEFTIADGD 60
M++ R++G V+WFN+ KGFGFI PDD +DLFVHQS+I+SEGFR L EG+ VEF +A
Sbjct: 33 MSDSRVNGKVKWFNDQKGFGFITPDDGGDDLFVHQSEIKSEGFRCLGEGEDVEFIVASDP 212
Query: 61 NGKTKAVDVTGPKGEPLQVRQDNHGGGGGGRGFRGGERRNGGGGCYTCGDTGHIARDCDR 120
+G+ KA++VTGP G+ +Q + G GGGGRG GG GG G
Sbjct: 213 DGRAKAIEVTGPNGQAVQGTRRGGGAGGGGRG--GGGYGGGGYG---------------- 338
Query: 121 SDRNDRNDRSGGGGGGDRDRACYTCGSFEHFARDCMRGGGNNNNGGGGYGGGGTSCYRCG 180
GG GGG R Y GGG G GGYGGGG Y G
Sbjct: 339 ---------GGGYGGGGRGGGGY--------------GGGGGYGGSGGYGGGGG--YGGG 443
Query: 181 GVGHIARDCATPSSGGGGGG 200
G G GGGGGG
Sbjct: 444 GRG--------GGRGGGGGG 479
Score = 47.8 bits (112), Expect = 2e-06
Identities = 36/90 (40%), Positives = 41/90 (45%)
Frame = +3
Query: 166 GGGYGGGGTSCYRCGGVGHIARDCATPSSGGGGGGACYKCGEVGHIARDCSNEGGRFDGG 225
GGG GGGG GG G+ GGG GG Y G G GG + GG
Sbjct: 279 GGGAGGGGR-----GGGGY---------GGGGYGGGGYGGGGRG---------GGGYGGG 389
Query: 226 NGRYDDGNGRFGGGNRRFGSGGGGHDGGKG 255
G G+G +GGG G GGGG GG+G
Sbjct: 390 GGY--GGSGGYGGGG---GYGGGGRGGGRG 464
Score = 37.4 bits (85), Expect = 0.003
Identities = 27/80 (33%), Positives = 32/80 (39%), Gaps = 1/80 (1%)
Frame = +3
Query: 177 YRCGGVGHIARDCATPSSGGGGGGACYKCGEVGHIARDCSNEGGRFDGGNGRYDDGNGRF 236
+RC G G + S G A G G + GG GG G G G +
Sbjct: 159 FRCLGEGEDV-EFIVASDPDGRAKAIEVTGPNGQAVQGTRRGGGAGGGGRGGGGYGGGGY 335
Query: 237 GGGNRRFGS-GGGGHDGGKG 255
GGG G GGGG+ GG G
Sbjct: 336 GGGGYGGGGRGGGGYGGGGG 395
>TC11885 similar to UP|Q8LF59 (Q8LF59) DNA-binding protein, partial (26%)
Length = 555
Score = 54.3 bits (129), Expect(2) = 1e-09
Identities = 35/103 (33%), Positives = 41/103 (38%), Gaps = 3/103 (2%)
Frame = +2
Query: 120 RSDRNDRND---RSGGGGGGDRDRACYTCGSFEHFARDCMRGGGNNNNGGGGYGGGGTSC 176
RSDR D R G RD C C HFAR+C C
Sbjct: 269 RSDRFSYRDAPYRRDSRRGFSRDNLCKNCKRPGHFAREC---------------PNVAIC 403
Query: 177 YRCGGVGHIARDCATPSSGGGGGGACYKCGEVGHIARDCSNEG 219
+ CG GHIA +C T S C+ C E GH+A C N+G
Sbjct: 404 HNCGLPGHIASECTTKS-------LCWNCKEPGHMASSCPNDG 511
Score = 27.7 bits (60), Expect = 2.2
Identities = 9/14 (64%), Positives = 11/14 (78%)
Frame = +2
Query: 257 CFNCGKAGHFARDC 270
C NC + GHFAR+C
Sbjct: 344 CKNCKRPGHFAREC 385
Score = 26.6 bits (57), Expect = 4.9
Identities = 17/73 (23%), Positives = 19/73 (25%)
Frame = +2
Query: 202 CYKCGEVGHIARDCSNEGGRFDGGNGRYDDGNGRFGGGNRRFGSGGGGHDGGKGTCFNCG 261
C C GH AR+C N C NCG
Sbjct: 344 CKNCKRPGHFARECPNVA------------------------------------ICHNCG 415
Query: 262 KAGHFARDCVEAS 274
GH A +C S
Sbjct: 416 LPGHIASECTTKS 454
Score = 26.2 bits (56), Expect(2) = 0.21
Identities = 14/55 (25%), Positives = 17/55 (30%)
Frame = +2
Query: 105 CYTCGDTGHIARDCDRSDRNDRNDRSGGGGGGDRDRACYTCGSFEHFARDCMRGG 159
C+ CG GHIA +C C+ C H A C G
Sbjct: 401 CHNCGLPGHIASECTTKS------------------LCWNCKEPGHMASSCPNDG 511
Score = 24.3 bits (51), Expect(2) = 1e-09
Identities = 8/11 (72%), Positives = 9/11 (81%)
Frame = +1
Query: 260 CGKAGHFARDC 270
CGK GH AR+C
Sbjct: 523 CGKVGHRAREC 555
Score = 23.5 bits (49), Expect(2) = 0.21
Identities = 8/13 (61%), Positives = 10/13 (76%)
Frame = +1
Query: 177 YRCGGVGHIARDC 189
+ CG VGH AR+C
Sbjct: 517 HTCGKVGHRAREC 555
>TC14758 similar to UP|O24601 (O24601) Glycine-rich RNA binding protein 2,
partial (93%)
Length = 725
Score = 58.5 bits (140), Expect = 1e-09
Identities = 45/129 (34%), Positives = 50/129 (37%)
Frame = +1
Query: 124 NDRNDRSGGGGGGDRDRACYTCGSFEHFARDCMRGGGNNNNGGGGYGGGGTSCYRCGGVG 183
N+ R GGGGGG R Y GGG GG GGGG+ Y GG G
Sbjct: 304 NEAQARGGGGGGGGRGGGGY--------------GGGGGRREGGYGGGGGSRGYGGGGGG 441
Query: 184 HIARDCATPSSGGGGGGACYKCGEVGHIARDCSNEGGRFDGGNGRYDDGNGRFGGGNRRF 243
+ GGGGG Y GGR +GG G GG R+
Sbjct: 442 Y-----------GGGGGGGY---------------GGRREGGYGG--------DGGGSRY 519
Query: 244 GSGGGGHDG 252
GSGGGG G
Sbjct: 520 GSGGGGGGG 546
Score = 56.2 bits (134), Expect = 6e-09
Identities = 48/149 (32%), Positives = 55/149 (36%)
Frame = +1
Query: 52 VEFTIADGDNGKTKAVDVTGPKGEPLQVRQDNHGGGGGGRGFRGGERRNGGGGCYTCGDT 111
V FT + + ++ G + V + GGGGG G RGG GGGG
Sbjct: 220 VTFTSEEAMRSAIEGMNGNELDGRNITVNEAQARGGGGGGGGRGGGGYGGGGG------- 378
Query: 112 GHIARDCDRSDRNDRNDRSGGGGGGDRDRACYTCGSFEHFARDCMRGGGNNNNGGGGYGG 171
R + GGGGG R Y G GGG GGGGYGG
Sbjct: 379 --------------RREGGYGGGGGSRG---YGGG-----------GGGYGGGGGGGYGG 474
Query: 172 GGTSCYRCGGVGHIARDCATPSSGGGGGG 200
R GG G S GGGGGG
Sbjct: 475 -----RREGGYGGDGGGSRYGSGGGGGGG 546
Score = 49.3 bits (116), Expect = 7e-07
Identities = 28/75 (37%), Positives = 36/75 (47%)
Frame = +1
Query: 195 GGGGGGACYKCGEVGHIARDCSNEGGRFDGGNGRYDDGNGRFGGGNRRFGSGGGGHDGGK 254
GGGGGG GG + GG GR + G G GGG+R +G GGGG+ GG
Sbjct: 322 GGGGGGG--------------GRGGGGYGGGGGRREGGYGG-GGGSRGYGGGGGGYGGGG 456
Query: 255 GTCFNCGKAGHFARD 269
G + + G + D
Sbjct: 457 GGGYGGRREGGYGGD 501
Score = 42.0 bits (97), Expect = 1e-04
Identities = 26/66 (39%), Positives = 30/66 (45%)
Frame = +1
Query: 80 RQDNHGGGGGGRGFRGGERRNGGGGCYTCGDTGHIARDCDRSDRNDRNDRSGGGGGGDRD 139
R+ +GGGGG RG+ GG GGGG G G D SGGGGGG
Sbjct: 382 REGGYGGGGGSRGYGGGGGGYGGGG---GGGYGGRREGGYGGDGGGSRYGSGGGGGGGNW 552
Query: 140 RACYTC 145
R+ C
Sbjct: 553 RS*IVC 570
Score = 31.2 bits (69), Expect = 0.20
Identities = 16/37 (43%), Positives = 19/37 (51%)
Frame = +1
Query: 219 GGRFDGGNGRYDDGNGRFGGGNRRFGSGGGGHDGGKG 255
G DG N ++ R GGG G GGGG+ GG G
Sbjct: 271 GNELDGRNITVNEAQAR-GGGGGGGGRGGGGYGGGGG 378
>TC11095 similar to PIR|T05112|T05112 splicing factor 9G8-like SR protein
RSZp22 [validated] - Arabidopsis thaliana
{Arabidopsis thaliana;}, partial (89%)
Length = 912
Score = 57.0 bits (136), Expect = 3e-09
Identities = 34/71 (47%), Positives = 40/71 (55%), Gaps = 3/71 (4%)
Frame = +1
Query: 161 NNNNGGGGYGGGGTSCYRCGGVGHIARDCATPSSGGGGGGA---CYKCGEVGHIARDCSN 217
+N+ GGGG GGGG R GG SGGGGGG+ CY+CGE GH AR+C
Sbjct: 292 HNSRGGGGGGGGG----RGGG-----------RSGGGGGGSDMKCYECGEPGHFAREC-- 420
Query: 218 EGGRFDGGNGR 228
R GG+GR
Sbjct: 421 ---RMRGGSGR 444
Score = 54.3 bits (129), Expect = 2e-08
Identities = 34/93 (36%), Positives = 42/93 (44%), Gaps = 1/93 (1%)
Frame = +1
Query: 68 DVTGPKGEPLQVRQDNHGGGGGGRGFRGGERRNGGGGCYTCGDTGHIARDCDRSDRNDRN 127
++ G G +++ ++ GGGGGG G RGG
Sbjct: 253 ELDGKNGWRVELSHNSRGGGGGGGGGRGG------------------------------- 339
Query: 128 DRSGGGGGGDRDRACYTCGSFEHFARDC-MRGG 159
RSGGGGGG D CY CG HFAR+C MRGG
Sbjct: 340 GRSGGGGGGS-DMKCYECGEPGHFARECRMRGG 435
Score = 50.4 bits (119), Expect = 3e-07
Identities = 26/64 (40%), Positives = 30/64 (46%), Gaps = 6/64 (9%)
Frame = +1
Query: 213 RDCSNEGGRFDGGNG------RYDDGNGRFGGGNRRFGSGGGGHDGGKGTCFNCGKAGHF 266
RD + DG NG G G GGG R G GGG G C+ CG+ GHF
Sbjct: 229 RDAQDAIRELDGKNGWRVELSHNSRGGGGGGGGGRGGGRSGGGGGGSDMKCYECGEPGHF 408
Query: 267 ARDC 270
AR+C
Sbjct: 409 AREC 420
Score = 48.1 bits (113), Expect = 2e-06
Identities = 27/70 (38%), Positives = 29/70 (40%)
Frame = +1
Query: 127 NDRSGGGGGGDRDRACYTCGSFEHFARDCMRGGGNNNNGGGGYGGGGTSCYRCGGVGHIA 186
N R GGGGGG RGGG + GGG GG CY CG GH A
Sbjct: 295 NSRGGGGGGGGG------------------RGGGRS---GGGGGGSDMKCYECGEPGHFA 411
Query: 187 RDCATPSSGG 196
R+C G
Sbjct: 412 RECRMRGGSG 441
Score = 37.7 bits (86), Expect = 0.002
Identities = 21/57 (36%), Positives = 25/57 (43%)
Frame = +1
Query: 85 GGGGGGRGFRGGERRNGGGGCYTCGDTGHIARDCDRSDRNDRNDRSGGGGGGDRDRA 141
GGGGGG + CY CG+ GH AR+C R GG G R R+
Sbjct: 349 GGGGGGSDMK----------CYECGEPGHFAREC----------RMRGGSGRRRSRS 459
>BP079571
Length = 414
Score = 56.6 bits (135), Expect = 4e-09
Identities = 38/93 (40%), Positives = 45/93 (47%), Gaps = 3/93 (3%)
Frame = -1
Query: 186 ARDCATP---SSGGGGGGACYKCGEVGHIARDCSNEGGRFDGGNGRYDDGNGRFGGGNRR 242
ARDCATP S+GG GGG C++ GEVGH+ARDC DGG
Sbjct: 414 ARDCATPKGPSNGGVGGGGCFRFGEVGHLARDC-------DGGVA--------------- 301
Query: 243 FGSGGGGHDGGKGTCFNCGKAGHFARDCVEASG 275
+ K T G G FAR+C+EASG
Sbjct: 300 -VTWWWWECWEKHTASIVGSLGTFARECLEASG 205
Score = 46.2 bits (108), Expect = 6e-06
Identities = 32/85 (37%), Positives = 41/85 (47%), Gaps = 11/85 (12%)
Frame = -1
Query: 152 ARDCMRGGGNNNNGGGGYGGGGTSCYRCGGVGHIARDCATPSSGGGGGGACYK------C 205
ARDC G +N GG GGGG C+R G VGH+ARDC + C++
Sbjct: 414 ARDCATPKGPSN---GGVGGGG--CFRFGEVGHLARDCDGGVAVTWWWWECWEKHTASIV 250
Query: 206 GEVGHIARDCSNEGG-----RFDGG 225
G +G AR+C G RF+ G
Sbjct: 249 GSLGTFARECLEASG*KCT*RFEEG 175
Score = 38.1 bits (87), Expect = 0.002
Identities = 13/19 (68%), Positives = 16/19 (83%)
Frame = -1
Query: 101 GGGGCYTCGDTGHIARDCD 119
GGGGC+ G+ GH+ARDCD
Sbjct: 369 GGGGCFRFGEVGHLARDCD 313
>TC15523 similar to UP|GRPA_MAIZE (P10979) Glycine-rich RNA-binding,
abscisic acid-inducible protein, complete
Length = 972
Score = 55.5 bits (132), Expect = 1e-08
Identities = 39/97 (40%), Positives = 41/97 (42%), Gaps = 3/97 (3%)
Frame = +2
Query: 158 GGGNNNNGG---GGYGGGGTSCYRCGGVGHIARDCATPSSGGGGGGACYKCGEVGHIARD 214
GGG GG GGYGGGG S Y GG G GG GGG
Sbjct: 611 GGGGGYGGGRREGGYGGGGGSRYSGGGGG---------GYGGSGGG-------------- 721
Query: 215 CSNEGGRFDGGNGRYDDGNGRFGGGNRRFGSGGGGHD 251
GGR +GG G Y G R GGG R+ SGG D
Sbjct: 722 --GYGGRREGGGGGY--GGSREGGGGSRYSSGGSWRD 820
Score = 50.4 bits (119), Expect = 3e-07
Identities = 45/148 (30%), Positives = 53/148 (35%), Gaps = 5/148 (3%)
Frame = +2
Query: 39 RSEGFRSLSEGDRVEFTIA-DGDNGKTKAVDVTGPKGEPLQVRQDNHGGGGGGRGFRGGE 97
RS GF ++ A +G NG+ D+ G + + GGGGGG G+ GG
Sbjct: 473 RSRGFGFVTFASEQSMKDAIEGMNGQ----DIDGRNVTVNEAQARGRGGGGGGGGYGGGR 640
Query: 98 RRNGGGGCYTCGDTGHIARDCDRSDRNDRNDRSGGGGGGDRDRACYTCGSFEHFARDCMR 157
R G GG R GGGG G Y
Sbjct: 641 REGGYGG-----------------GGGSRYSGGGGGGYGGSGGGGY-------------- 727
Query: 158 GGGNNNNGGGGYG----GGGTSCYRCGG 181
GG GGGGYG GGG S Y GG
Sbjct: 728 -GGRREGGGGGYGGSREGGGGSRYSSGG 808
Score = 45.4 bits (106), Expect = 1e-05
Identities = 36/108 (33%), Positives = 42/108 (38%), Gaps = 3/108 (2%)
Frame = +2
Query: 157 RGGGNNNNGGGGYGGGGTSCYRCGGVGHIARDCATPSSGGGGGGACYKCGEVGHIARDCS 216
RGGG GGGGYGGG GG GGG
Sbjct: 599 RGGGG---GGGGYGGGRRE-------------------GGYGGGG--------------- 667
Query: 217 NEGGRFDGGNGRYDDGNGRFGGGNRRFGSG---GGGHDGGKGTCFNCG 261
G R+ GG G G+G G G RR G G GG +GG G+ ++ G
Sbjct: 668 --GSRYSGGGGGGYGGSGGGGYGGRREGGGGGYGGSREGGGGSRYSSG 805
Score = 28.5 bits (62), Expect = 1.3
Identities = 16/43 (37%), Positives = 22/43 (50%)
Frame = +2
Query: 224 GGNGRYDDGNGRFGGGNRRFGSGGGGHDGGKGTCFNCGKAGHF 266
GG G G G +GGG R GG+ GG G+ ++ G G +
Sbjct: 602 GGGG----GGGGYGGGRRE-----GGYGGGGGSRYSGGGGGGY 703
>TC9521 similar to UP|Q9FYA7 (Q9FYA7) Splicing factor RSZ33, partial (56%)
Length = 598
Score = 53.9 bits (128), Expect = 3e-08
Identities = 34/101 (33%), Positives = 42/101 (40%), Gaps = 4/101 (3%)
Frame = +1
Query: 121 SDRNDRNDRSGGGGGGDRDRACYTCGSFEHFARDCMRGGGNNNNG----GGGYGGGGTSC 176
SD D +D G D D + + R GGG + G G G C
Sbjct: 214 SDPRDADDARYNLDGRDVDGSRLIVEFAKGVPRGSREGGGGRDRDREYMGRGPPPGSGRC 393
Query: 177 YRCGGVGHIARDCATPSSGGGGGGACYKCGEVGHIARDCSN 217
+ CG GH ARDC G CY+CGE GHI ++C N
Sbjct: 394 FNCGIDGHWARDC----KAGDWKNKCYRCGERGHIEKNCKN 504
Score = 47.0 bits (110), Expect = 4e-06
Identities = 41/140 (29%), Positives = 53/140 (37%), Gaps = 3/140 (2%)
Frame = +1
Query: 19 FGFIKPDDDSEDL-FVHQSDIRSEGFRSLSEGDRVEFTIADGDNGKTKAVDVTGPKGEPL 77
+G ++ D D FV SD R + D + + DG + + V KG P
Sbjct: 160 YGRVRDVDMKRDYAFVEFSDPR--------DADDARYNL-DGRDVDGSRLIVEFAKGVP- 309
Query: 78 QVRQDNHGGGGGGRG--FRGGERRNGGGGCYTCGDTGHIARDCDRSDRNDRNDRSGGGGG 135
R GGGG R + G G G C+ CG GH ARDC
Sbjct: 310 --RGSREGGGGRDRDREYMGRGPPPGSGRCFNCGIDGHWARDCK---------------A 438
Query: 136 GDRDRACYTCGSFEHFARDC 155
GD CY CG H ++C
Sbjct: 439 GDWKNKCYRCGERGHIEKNC 498
Score = 39.3 bits (90), Expect = 7e-04
Identities = 18/34 (52%), Positives = 20/34 (57%)
Frame = +1
Query: 237 GGGNRRFGSGGGGHDGGKGTCFNCGKAGHFARDC 270
GG +R G G G G CFNCG GH+ARDC
Sbjct: 331 GGRDRDREYMGRGPPPGSGRCFNCGIDGHWARDC 432
Score = 35.0 bits (79), Expect = 0.014
Identities = 21/62 (33%), Positives = 26/62 (41%), Gaps = 11/62 (17%)
Frame = +1
Query: 220 GRFDGGNGRYDD----GNGRFGGGNRRFGSGGGGH-------DGGKGTCFNCGKAGHFAR 268
G +GG GR D G G G R F G GH K C+ CG+ GH +
Sbjct: 313 GSREGGGGRDRDREYMGRGPPPGSGRCFNCGIDGHWARDCKAGDWKNKCYRCGERGHIEK 492
Query: 269 DC 270
+C
Sbjct: 493 NC 498
Score = 33.1 bits (74), Expect = 0.053
Identities = 23/75 (30%), Positives = 35/75 (46%), Gaps = 2/75 (2%)
Frame = +1
Query: 58 DGDNGKTKAVDVTGPKGEPLQVRQDNHG-GGGGGRGFRGGERRNGGGGCYTCGDTGHIAR 116
+G G+ + + G P R N G G R + G+ +N CY CG+ GHI +
Sbjct: 322 EGGGGRDRDREYMGRGPPPGSGRCFNCGIDGHWARDCKAGDWKNK---CYRCGERGHIEK 492
Query: 117 DCDRSDRN-DRNDRS 130
+C S + R+ RS
Sbjct: 493 NCKNSPKKLSRHPRS 537
>TC16065 weakly similar to UP|Q9SBM1 (Q9SBM1) Hydroxyproline-rich
glycoprotein DZ-HRGP precursor, partial (20%)
Length = 608
Score = 51.6 bits (122), Expect = 1e-07
Identities = 51/166 (30%), Positives = 61/166 (36%), Gaps = 2/166 (1%)
Frame = -1
Query: 92 GFRGGERRNGGGGCYTCGDTGHIARDCDRSDRNDRNDRSGGGGGGDRDRACYTCGSFEHF 151
GF +R G G G + +RS + + GGGG + G
Sbjct: 494 GFIMVRKRRTDGPSGGEGSEGQHSHSTERSAPPPQQAAAAPGGGGPGPQGGRGYGGPPQG 315
Query: 152 ARDCMRGGGNNNNGGGGYGGGGTSCYRCGGVGHIARDCATPS--SGGGGGGACYKCGEVG 209
R GGG GG GYGGGG GG G + P G G GG
Sbjct: 314 GR----GGGYGGGGGPGYGGGGR-----GGHGVPQQQYGGPPEYQGRGRGGP-------- 186
Query: 210 HIARDCSNEGGRFDGGNGRYDDGNGRFGGGNRRFGSGGGGHDGGKG 255
S +GGR G Y G G +GGG G GGGG G+G
Sbjct: 185 ------SQQGGR-----GGYSGGGGGYGGGG---GRGGGGMGSGRG 90
Score = 42.7 bits (99), Expect = 7e-05
Identities = 42/123 (34%), Positives = 49/123 (39%), Gaps = 11/123 (8%)
Frame = -1
Query: 85 GGGG----GGRGFRGGERRNGGGGCYTCGDTGHIARDCDRSDRNDRNDRSGGGGGGDRDR 140
GGGG GGRG+ G + GGG G G+ GGG GG
Sbjct: 371 GGGGPGPQGGRGYGGPPQGGRGGGYGGGGGPGY----------------GGGGRGGHGVP 240
Query: 141 ACYTCGSFEHFARDCMRGG-------GNNNNGGGGYGGGGTSCYRCGGVGHIARDCATPS 193
G E+ R RGG G + GGGGYGGGG GG G + +PS
Sbjct: 239 QQQYGGPPEYQGRG--RGGPSQQGGRGGYSGGGGGYGGGGGR----GGGGMGSGRGVSPS 78
Query: 194 SGG 196
GG
Sbjct: 77 HGG 69
Score = 28.1 bits (61), Expect = 1.7
Identities = 16/32 (50%), Positives = 18/32 (56%), Gaps = 1/32 (3%)
Frame = -1
Query: 73 KGEPLQVRQDNHGG-GGGGRGFRGGERRNGGG 103
+G P Q Q GG GGG G+ GG R GGG
Sbjct: 197 RGGPSQ--QGGRGGYSGGGGGYGGGGGRGGGG 108
>TC10011 similar to UP|Q9FYA7 (Q9FYA7) Splicing factor RSZ33, partial (62%)
Length = 684
Score = 51.6 bits (122), Expect = 1e-07
Identities = 26/63 (41%), Positives = 32/63 (50%), Gaps = 2/63 (3%)
Frame = +2
Query: 157 RGG--GNNNNGGGGYGGGGTSCYRCGGVGHIARDCATPSSGGGGGGACYKCGEVGHIARD 214
+GG GN G G G C+ CG GH ARDC G CY+CG+ GH+ R+
Sbjct: 308 KGGPRGNREYLGRGPPPGSGRCFNCGLDGHWARDC----KAGDWKNKCYRCGDRGHVERN 475
Query: 215 CSN 217
C N
Sbjct: 476 CKN 484
Score = 46.2 bits (108), Expect = 6e-06
Identities = 26/78 (33%), Positives = 29/78 (36%)
Frame = +2
Query: 85 GGGGGGRGFRGGERRNGGGGCYTCGDTGHIARDCDRSDRNDRNDRSGGGGGGDRDRACYT 144
GG G R + G G G C+ CG GH ARDC GD CY
Sbjct: 311 GGPRGNREYLGRGPPPGSGRCFNCGLDGHWARDCK---------------AGDWKNKCYR 445
Query: 145 CGSFEHFARDCMRGGGNN 162
CG H R+C N
Sbjct: 446 CGDRGHVERNCKNSPKKN 499
Score = 39.3 bits (90), Expect = 7e-04
Identities = 18/32 (56%), Positives = 20/32 (62%)
Frame = +2
Query: 239 GNRRFGSGGGGHDGGKGTCFNCGKAGHFARDC 270
GNR + G G G G CFNCG GH+ARDC
Sbjct: 323 GNREYL--GRGPPPGSGRCFNCGLDGHWARDC 412
Score = 33.1 bits (74), Expect = 0.053
Identities = 20/73 (27%), Positives = 25/73 (33%)
Frame = +2
Query: 198 GGGACYKCGEVGHIARDCSNEGGRFDGGNGRYDDGNGRFGGGNRRFGSGGGGHDGGKGTC 257
G G C+ CG GH ARDC + G + K C
Sbjct: 359 GSGRCFNCGLDGHWARDC--KAGDW-------------------------------KNKC 439
Query: 258 FNCGKAGHFARDC 270
+ CG GH R+C
Sbjct: 440 YRCGDRGHVERNC 478
>AV773507
Length = 496
Score = 49.7 bits (117), Expect = 5e-07
Identities = 35/94 (37%), Positives = 41/94 (43%), Gaps = 2/94 (2%)
Frame = +2
Query: 158 GGGNNNNG--GGGYGGGGTSCYRCGGVGHIARDCATPSSGGGGGGACYKCGEVGHIARDC 215
GG + + G GGGY GG Y G + +D C++CG GH ARDC
Sbjct: 254 GGDDADQGYRGGGYSSGGRGSYGAGD--RVGQD------------DCFECGRPGHRARDC 391
Query: 216 SNEGGRFDGGNGRYDDGNGRFGGGNRRFGSGGGG 249
GG GG GR G G R GSGG G
Sbjct: 392 PLAGG--GGGRGR---GGGSLSSRPRYGGSGGHG 478
Score = 49.3 bits (116), Expect = 7e-07
Identities = 24/47 (51%), Positives = 28/47 (59%), Gaps = 4/47 (8%)
Frame = +2
Query: 157 RGGGNNNNGGGGYGGGGT----SCYRCGGVGHIARDCATPSSGGGGG 199
RGGG ++ G G YG G C+ CG GH ARDC P +GGGGG
Sbjct: 281 RGGGYSSGGRGSYGAGDRVGQDDCFECGRPGHRARDC--PLAGGGGG 415
Score = 45.8 bits (107), Expect = 8e-06
Identities = 38/118 (32%), Positives = 44/118 (37%), Gaps = 2/118 (1%)
Frame = +2
Query: 66 AVDVTGPKGEPLQVRQDNHGGG--GGGRGFRGGERRNGGGGCYTCGDTGHIARDCDRSDR 123
+V+ PK Q GGG GGRG G R G C+ CG GH ARDC
Sbjct: 227 SVNKAQPKMGGDDADQGYRGGGYSSGGRGSYGAGDRVGQDDCFECGRPGHRARDC----- 391
Query: 124 NDRNDRSGGGGGGDRDRACYTCGSFEHFARDCMRGGGNNNNGGGGYGGGGTSCYRCGG 181
+GGGGG R GG + + YGG G R GG
Sbjct: 392 ----PLAGGGGGRGR--------------------GGGSLSSRPRYGGSGGHGDRLGG 493
Score = 41.6 bits (96), Expect = 1e-04
Identities = 23/51 (45%), Positives = 25/51 (48%), Gaps = 2/51 (3%)
Frame = +2
Query: 227 GRYDDGNGRFGGGNRRFGSG--GGGHDGGKGTCFNCGKAGHFARDCVEASG 275
G D G GGG G G G G G+ CF CG+ GH ARDC A G
Sbjct: 254 GGDDADQGYRGGGYSSGGRGSYGAGDRVGQDDCFECGRPGHRARDCPLAGG 406
>TC18003 similar to PIR|T05112|T05112 splicing factor 9G8-like SR protein
RSZp22 [validated] - Arabidopsis thaliana
{Arabidopsis thaliana;}, partial (42%)
Length = 587
Score = 46.2 bits (108), Expect = 6e-06
Identities = 28/75 (37%), Positives = 36/75 (47%), Gaps = 2/75 (2%)
Frame = +1
Query: 71 GPKGEPLQVRQDNHGGGGGGRGFRGGERRNGGGG--CYTCGDTGHIARDCDRSDRNDRND 128
G G +++ ++ GGGGG RGG R GG CY CG+ GH AR+C R
Sbjct: 70 GKNGWRVELSHNSKGGGGG----RGGGRGRGGEDLKCYECGEPGHFAREC-----RSRGG 222
Query: 129 RSGGGGGGDRDRACY 143
G G G R + Y
Sbjct: 223 SRGLGSGRRRSPSPY 267
Score = 46.2 bits (108), Expect = 6e-06
Identities = 21/43 (48%), Positives = 23/43 (52%)
Frame = +1
Query: 157 RGGGNNNNGGGGYGGGGTSCYRCGGVGHIARDCATPSSGGGGG 199
+GGG GG G GG CY CG GH AR+C S GG G
Sbjct: 109 KGGGGGRGGGRGRGGEDLKCYECGEPGHFARECR--SRGGSRG 231
Score = 45.8 bits (107), Expect = 8e-06
Identities = 25/68 (36%), Positives = 30/68 (43%)
Frame = +1
Query: 161 NNNNGGGGYGGGGTSCYRCGGVGHIARDCATPSSGGGGGGACYKCGEVGHIARDCSNEGG 220
N+ GGGG GGG GG CY+CGE GH AR+C + GG
Sbjct: 103 NSKGGGGGRGGG--------------------RGRGGEDLKCYECGEPGHFARECRSRGG 222
Query: 221 RFDGGNGR 228
G+GR
Sbjct: 223 SRGLGSGR 246
Score = 44.3 bits (103), Expect = 2e-05
Identities = 19/39 (48%), Positives = 21/39 (53%)
Frame = +1
Query: 129 RSGGGGGGDRDRACYTCGSFEHFARDCMRGGGNNNNGGG 167
R GG G G D CY CG HFAR+C GG+ G G
Sbjct: 127 RGGGRGRGGEDLKCYECGEPGHFARECRSRGGSRGLGSG 243
Score = 43.9 bits (102), Expect = 3e-05
Identities = 25/55 (45%), Positives = 30/55 (54%), Gaps = 2/55 (3%)
Frame = +1
Query: 223 DGGNG-RYD-DGNGRFGGGNRRFGSGGGGHDGGKGTCFNCGKAGHFARDCVEASG 275
DG NG R + N + GGG R G G GG D C+ CG+ GHFAR+C G
Sbjct: 67 DGKNGWRVELSHNSKGGGGGRGGGRGRGGEDL---KCYECGEPGHFARECRSRGG 222
>TC19911 weakly similar to PIR|A44805|A44805 eggshell protein precursor -
fluke (Schistosoma haematobium) {Schistosoma
haematobium;} , partial (24%)
Length = 350
Score = 46.2 bits (108), Expect = 6e-06
Identities = 40/123 (32%), Positives = 45/123 (36%)
Frame = +1
Query: 130 SGGGGGGDRDRACYTCGSFEHFARDCMRGGGNNNNGGGGYGGGGTSCYRCGGVGHIARDC 189
SGGG G+ +H D GG + G G GGG D
Sbjct: 40 SGGGSHGN-----------DHRHSDNEHGGRRSETTGHGLSGGGY-------------DD 147
Query: 190 ATPSSGGGGGGACYKCGEVGHIARDCSNEGGRFDGGNGRYDDGNGRFGGGNRRFGSGGGG 249
S G GGG G+ GH A + GGR GG G Y DG G G G G GG
Sbjct: 148 DNRSGGRTGGGY----GDGGHSAGGYGDHGGRSGGGGGGYGDGGLSSGSG----GYGDGG 303
Query: 250 HDG 252
G
Sbjct: 304 RSG 312
Score = 44.7 bits (104), Expect = 2e-05
Identities = 39/117 (33%), Positives = 46/117 (38%), Gaps = 1/117 (0%)
Frame = +1
Query: 85 GGGGGGRGFRGGERRNGGGGCYTCGDTGHIARDCDRSDRNDRNDRSGGGGGGDRDRACYT 144
GGG G R + +GG T TGH D N RSGG GG ++
Sbjct: 43 GGGSHGNDHRHSDNEHGGRRSET---TGHGLSGGGYDDDN----RSGGRTGGGYGDGGHS 201
Query: 145 CGSF-EHFARDCMRGGGNNNNGGGGYGGGGTSCYRCGGVGHIARDCATPSSGGGGGG 200
G + +H GG + GGGGYG GG S GG G R G GG
Sbjct: 202 AGGYGDH--------GGRSGGGGGGYGDGGLSS-GSGGYGDGGRSGVGYGDGXLSGG 345
Score = 38.5 bits (88), Expect = 0.001
Identities = 35/112 (31%), Positives = 43/112 (38%), Gaps = 8/112 (7%)
Frame = +1
Query: 162 NNNGGGGYGGGGT--SCYRCGGVGHIARDCATPSSGGGGGGACYKCGEVGHIARDCSNEG 219
N+ G G+ GGG+ + +R H R T G GGG G +G
Sbjct: 16 NSAGNTGHSGGGSHGNDHRHSDNEHGGRRSETTGHGLSGGGYDDDNRSGGRTGGGYG-DG 192
Query: 220 GRFDGG----NGRYDDGNGRFGGGNRRFGSGGGGHDGGKGTCFNCG--KAGH 265
G GG GR G G +G G GSGG G G G + G GH
Sbjct: 193 GHSAGGYGDHGGRSGGGGGGYGDGGLSSGSGGYGDGGRSGVGYGDGXLSGGH 348
>TC19964 similar to UP|Q40363 (Q40363) NuM1 protein, partial (11%)
Length = 562
Score = 45.8 bits (107), Expect = 8e-06
Identities = 38/90 (42%), Positives = 41/90 (45%), Gaps = 7/90 (7%)
Frame = +2
Query: 185 IARDCATPSSGGGGGGACYKCGEVGHIARDCSNEGGRFDGGNGRYDDGNGRFGGGNR--- 241
+A D A P G GG + G R GGRFD G G G GRFG G R
Sbjct: 101 LAVDEAKPRDNQGSGG---RSGG----GRSGGGRGGRFDSGRG---GGRGRFGSGGRGGD 250
Query: 242 ---RFGSGGGGHDGGKGTCFN-CGKAGHFA 267
R GSGGGG GG+GT GK FA
Sbjct: 251 RGGRGGSGGGGR-GGRGTPLRAAGKKTTFA 337
Score = 41.6 bits (96), Expect = 1e-04
Identities = 36/125 (28%), Positives = 45/125 (35%), Gaps = 3/125 (2%)
Frame = +2
Query: 15 NTKGFGFIKPDDDS---EDLFVHQSDIRSEGFRSLSEGDRVEFTIADGDNGKTKAVDVTG 71
N KGF ++ D + + L +H SD+ G T AVD
Sbjct: 8 NXKGFAYLDFSDTNSVNKALELHDSDL----------------------GGFTLAVDEAK 121
Query: 72 PKGEPLQVRQDNHGGGGGGRGFRGGERRNGGGGCYTCGDTGHIARDCDRSDRNDRNDRSG 131
P+ + G GGGRG R R GG G + G G DR R G
Sbjct: 122 PRDNQGSGGRSGGGRSGGGRGGRFDSGRGGGRGRFGSGGRG-----------GDRGGRGG 268
Query: 132 GGGGG 136
GGGG
Sbjct: 269 SGGGG 283
>AV427333
Length = 387
Score = 45.4 bits (106), Expect = 1e-05
Identities = 45/127 (35%), Positives = 48/127 (37%), Gaps = 7/127 (5%)
Frame = -3
Query: 85 GGGGGGRGFRGGERRNGGGGCYTCGDTGHIARDCDRSDRNDRNDRSGGGGGGDRDRACYT 144
G G GG G GG R GGG CGD G R C N +D GG GG
Sbjct: 370 GEGEGGSG-GGGGRTCGGGVIG*CGDGG--GRTCGGGGVNG*DDGGGGDDGG-------- 224
Query: 145 CGSFEHFARDCMRGGGNNNNGGGGYG-------GGGTSCYRCGGVGHIARDCATPSSGGG 197
C RGGG N GGG GGG + GGVG + G
Sbjct: 223 -------GEGCPRGGGANE*EGGGGDLK*WEGVGGGLKGWE-GGVGGL-------*GWEG 89
Query: 198 GGGACYK 204
GGG C K
Sbjct: 88 GGGKCGK 68
Score = 40.8 bits (94), Expect = 3e-04
Identities = 39/108 (36%), Positives = 43/108 (39%), Gaps = 2/108 (1%)
Frame = -3
Query: 93 FRGGERRNGGGGCYTCGDTGHIARDCDRSDRNDRNDRSGGGGGGDRDRACYTCGSFEHFA 152
F GE +GGGG TCG G + C G GGG TCG
Sbjct: 373 FGEGEGGSGGGGGRTCG--GGVIG*C------------GDGGG-------RTCG------ 275
Query: 153 RDCMRGGGNN--NNGGGGYGGGGTSCYRCGGVGHIARDCATPSSGGGG 198
GGG N ++GGGG GGG C R GG A GGGG
Sbjct: 274 -----GGGVNG*DDGGGGDDGGGEGCPRGGG--------ANE*EGGGG 170
Score = 40.4 bits (93), Expect = 3e-04
Identities = 27/65 (41%), Positives = 31/65 (47%), Gaps = 4/65 (6%)
Frame = -3
Query: 195 GGGGGGACYKCGEVGHIARDCSNEGGRFDGGNGRY--DDGNGRFGGGNRRFGSGGGGH-- 250
GG GGG CG G + C + GGR GG G DDG G GG GGG +
Sbjct: 358 GGSGGGGGRTCG--GGVIG*CGDGGGRTCGGGGVNG*DDGGGGDDGGGEGCPRGGGANE* 185
Query: 251 DGGKG 255
+GG G
Sbjct: 184 EGGGG 170
Score = 35.8 bits (81), Expect = 0.008
Identities = 32/88 (36%), Positives = 34/88 (38%), Gaps = 6/88 (6%)
Frame = -3
Query: 166 GGGYGGGGTSCYRCGGVGHIARDCATPSSGGGGGGACYKCGEVGHIARDCSNEG--GRFD 223
GG GGGG +C GGG CG+ G R C G G D
Sbjct: 358 GGSGGGGGRTC---------------------GGGVIG*CGDGG--GRTCGGGGVNG*DD 248
Query: 224 GGNGRYDDGNG----RFGGGNRRFGSGG 247
GG G DDG G R GG N G GG
Sbjct: 247 GGGG--DDGGGEGCPRGGGANE*EGGGG 170
>AV420911
Length = 418
Score = 44.7 bits (104), Expect = 2e-05
Identities = 22/50 (44%), Positives = 29/50 (58%), Gaps = 2/50 (4%)
Frame = +2
Query: 71 GPKGEPLQVRQDNHGGGGGGRGFRGGERRNGGGG--CYTCGDTGHIARDC 118
G G +++ ++ GGGGG RGG R GG CY CG+ GH AR+C
Sbjct: 278 GKNGWRVELSHNSKGGGGG----RGGGRGRGGEDLKCYECGEPGHFAREC 415
Score = 44.3 bits (103), Expect = 2e-05
Identities = 17/33 (51%), Positives = 19/33 (57%)
Frame = +2
Query: 157 RGGGNNNNGGGGYGGGGTSCYRCGGVGHIARDC 189
+GGG GG G GG CY CG GH AR+C
Sbjct: 317 KGGGGGRGGGRGRGGEDLKCYECGEPGHFAREC 415
Score = 43.9 bits (102), Expect = 3e-05
Identities = 26/60 (43%), Positives = 32/60 (53%), Gaps = 2/60 (3%)
Frame = +2
Query: 213 RDCSNEGGRFDGGNG-RYD-DGNGRFGGGNRRFGSGGGGHDGGKGTCFNCGKAGHFARDC 270
RD + DG NG R + N + GGG R G G GG D C+ CG+ GHFAR+C
Sbjct: 245 RDALDAIHALDGKNGWRVELSHNSKGGGGGRGGGRGRGGEDL---KCYECGEPGHFAREC 415
Score = 37.7 bits (86), Expect = 0.002
Identities = 20/55 (36%), Positives = 23/55 (41%)
Frame = +2
Query: 161 NNNNGGGGYGGGGTSCYRCGGVGHIARDCATPSSGGGGGGACYKCGEVGHIARDC 215
N+ GGGG GGG GG CY+CGE GH AR+C
Sbjct: 311 NSKGGGGGRGGG--------------------RGRGGEDLKCYECGEPGHFAREC 415
Score = 37.4 bits (85), Expect = 0.003
Identities = 15/27 (55%), Positives = 16/27 (58%)
Frame = +2
Query: 129 RSGGGGGGDRDRACYTCGSFEHFARDC 155
R GG G G D CY CG HFAR+C
Sbjct: 335 RGGGRGRGGEDLKCYECGEPGHFAREC 415
>TC17551 weakly similar to UP|AAR24662 (AAR24662) At5g26960, partial (22%)
Length = 505
Score = 44.3 bits (103), Expect = 2e-05
Identities = 35/95 (36%), Positives = 41/95 (42%)
Frame = -3
Query: 160 GNNNNGGGGYGGGGTSCYRCGGVGHIARDCATPSSGGGGGGACYKCGEVGHIARDCSNEG 219
G G G GG G S R R C S GG G G+ ++ G R
Sbjct: 440 GRWKRGARGRGGSGRSRGR--------RRC---SRGGRGRGSIWRRGGWRRRGRS----- 309
Query: 220 GRFDGGNGRYDDGNGRFGGGNRRFGSGGGGHDGGK 254
R+ G GR G G GGG R GSGGGG +GG+
Sbjct: 308 -RWR*GRGRSGGGGGGGGGGRRSRGSGGGGTNGGR 207
Score = 39.3 bits (90), Expect = 7e-04
Identities = 37/102 (36%), Positives = 42/102 (40%), Gaps = 6/102 (5%)
Frame = -1
Query: 158 GGGNNNNGGGGYGGGGTSCYRCGGVGHIARDCATPSSGGGGGGACYKCGEVGHIARD--C 215
GGG GGG G GG +A + A + G GG EVG A D
Sbjct: 331 GGGGEAEVGGGEGEGG-----------VAEEVAAAAEVGEVGGV----EEVGPTAADEGE 197
Query: 216 SNEGGRFDGGNGRYDD--GNGRFGGGN--RRFGSGGGGHDGG 253
EG +D G +D G GGG R FG GGGG GG
Sbjct: 196 GREGRGWDAGEEVENDVVREGGNGGGRLLRWFGDGGGGGGGG 71
Score = 33.1 bits (74), Expect = 0.053
Identities = 29/94 (30%), Positives = 33/94 (34%)
Frame = -3
Query: 90 GRGFRGGERRNGGGGCYTCGDTGHIARDCDRSDRNDRNDRSGGGGGGDRDRACYTCGSFE 149
GR RG R G G RS R R G G G + + G +
Sbjct: 440 GRWKRGARGRGGSG----------------RSRGRRRCSRGGRGRG-----SIWRRGGWR 324
Query: 150 HFARDCMRGGGNNNNGGGGYGGGGTSCYRCGGVG 183
R R G + GGGG GGGG GG G
Sbjct: 323 RRGRSRWR*GRGRSGGGGGGGGGGRRSRGSGGGG 222
Score = 30.0 bits (66), Expect = 0.45
Identities = 32/125 (25%), Positives = 46/125 (36%), Gaps = 9/125 (7%)
Frame = -1
Query: 57 ADGDNGKTKAVDVTGPKGEPLQVRQDNHGGGGGGRGFRGGERRNGGG-GCYTCGDTGHIA 115
ADG + V G +G+ + ++ G G GGE GGG G + A
Sbjct: 439 ADGSEARVGEEGVEGLEGDGVVAGEEEGGVPFGVEEGGGGEAEVGGGEGEGGVAEEVAAA 260
Query: 116 RDCDRSDRNDRNDRSGGGGGGDRDRACYTCGSFEHFARDCMRGGGNN--------NNGGG 167
+ + + G R+ + G E D +R GGN +GGG
Sbjct: 259 AEVGEVGGVEEVGPTAADEGEGREGRGWDAG--EEVENDVVREGGNGGGRLLRWFGDGGG 86
Query: 168 GYGGG 172
G GGG
Sbjct: 85 GGGGG 71
Score = 28.1 bits (61), Expect = 1.7
Identities = 24/81 (29%), Positives = 30/81 (36%), Gaps = 17/81 (20%)
Frame = -1
Query: 48 EGDRVEFTIADGDNGKTKAVDVTGPK---------------GEPLQVRQDNHGGGGGGRG 92
EG E A + G+ V+ GP GE ++ GG GGGR
Sbjct: 292 EGGVAEEVAAAAEVGEVGGVEEVGPTAADEGEGREGRGWDAGEEVENDVVREGGNGGGRL 113
Query: 93 FR--GGERRNGGGGCYTCGDT 111
R G GGGG G+T
Sbjct: 112 LRWFGDGGGGGGGGVVWIGET 50
>TC17853 similar to UP|Q42412 (Q42412) RNA-binding protein RZ-1, partial
(58%)
Length = 881
Score = 43.5 bits (101), Expect = 4e-05
Identities = 22/49 (44%), Positives = 26/49 (52%), Gaps = 1/49 (2%)
Frame = +2
Query: 223 DGGNGRYDDGNGRFGGGNRRFGSGGGGHDGGKG-TCFNCGKAGHFARDC 270
D G+ + G R G+R G GG G G CF CGK GHFAR+C
Sbjct: 314 DDGDRYRERGRDRDDRGDRDRSRGYGGSRGSNGGECFKCGKPGHFAREC 460
Score = 42.0 bits (97), Expect = 1e-04
Identities = 30/112 (26%), Positives = 52/112 (45%), Gaps = 2/112 (1%)
Frame = +2
Query: 9 VVQWFNNTKGFGFIKPDDDS--EDLFVHQSDIRSEGFRSLSEGDRVEFTIADGDNGKTKA 66
V ++ ++GFGF+ DD ++ + + +G + + + + D D+G
Sbjct: 155 VDKFSGRSRGFGFVTFDDKKAMDEAIDAMNGMDLDGRTITVDKAQPQGSSRDRDDG---- 322
Query: 67 VDVTGPKGEPLQVRQDNHGGGGGGRGFRGGERRNGGGGCYTCGDTGHIARDC 118
D +G +D+ G RG+ GG R + GG C+ CG GH AR+C
Sbjct: 323 -DRYRERGRD----RDDRGDRDRSRGY-GGSRGSNGGECFKCGKPGHFAREC 460
Score = 41.2 bits (95), Expect = 2e-04
Identities = 15/26 (57%), Positives = 19/26 (72%)
Frame = +2
Query: 194 SGGGGGGACYKCGEVGHIARDCSNEG 219
S G GG C+KCG+ GH AR+C +EG
Sbjct: 395 SRGSNGGECFKCGKPGHFARECPSEG 472
Score = 39.7 bits (91), Expect = 6e-04
Identities = 29/86 (33%), Positives = 35/86 (39%), Gaps = 5/86 (5%)
Frame = +2
Query: 115 ARDCDRSDR-----NDRNDRSGGGGGGDRDRACYTCGSFEHFARDCMRGGGNNNNGGGGY 169
+RD D DR DR+DR GDRDR+ GG
Sbjct: 302 SRDRDDGDRYRERGRDRDDR------GDRDRS---------------------RGYGGSR 400
Query: 170 GGGGTSCYRCGGVGHIARDCATPSSG 195
G G C++CG GH AR+C PS G
Sbjct: 401 GSNGGECFKCGKPGHFAREC--PSEG 472
Score = 30.4 bits (67), Expect = 0.34
Identities = 20/70 (28%), Positives = 29/70 (40%)
Frame = +3
Query: 66 AVDVTGPKGEPLQVRQDNHGGGGGGRGFRGGERRNGGGGCYTCGDTGHIARDCDRSDRND 125
+V V G +G +++ GGG R +R +GG G G D DR+
Sbjct: 456 SVLVKGKRGGGRYGGRESRXSGGGYGPDRNADRSSGGRSSRDGGSHGDSGSDRYHRDRSG 635
Query: 126 RNDRSGGGGG 135
+R G GG
Sbjct: 636 PYERRGSSGG 665
>BP030058
Length = 455
Score = 43.1 bits (100), Expect = 5e-05
Identities = 22/55 (40%), Positives = 25/55 (45%)
Frame = +1
Query: 195 GGGGGGACYKCGEVGHIARDCSNEGGRFDGGNGRYDDGNGRFGGGNRRFGSGGGG 249
GGGG CG+ G+ GG G G DG+G GGG G GGGG
Sbjct: 247 GGGGDKGLQFCGQEGYFGGGGEGGGGGGGDGGGGGGDGHGSGGGGGEGDGGGGGG 411
Score = 39.3 bits (90), Expect = 7e-04
Identities = 37/119 (31%), Positives = 43/119 (36%)
Frame = +1
Query: 133 GGGGDRDRACYTCGSFEHFARDCMRGGGNNNNGGGGYGGGGTSCYRCGGVGHIARDCATP 192
GGGGD+ CG +F GGG GGGG GGGG GG GH
Sbjct: 247 GGGGDK--GLQFCGQEGYFG-----GGGEGGGGGGGDGGGG------GGDGH-------- 363
Query: 193 SSGGGGGGACYKCGEVGHIARDCSNEGGRFDGGNGRYDDGNGRFGGGNRRFGSGGGGHD 251
SGGGGG +G+G GGG+ + GHD
Sbjct: 364 GSGGGGG-------------------------------EGDGGGGGGSPQVSP--QGHD 441
Score = 33.5 bits (75), Expect = 0.040
Identities = 22/66 (33%), Positives = 27/66 (40%)
Frame = +1
Query: 71 GPKGEPLQVRQDNHGGGGGGRGFRGGERRNGGGGCYTCGDTGHIARDCDRSDRNDRNDRS 130
G KG ++ GGGG G G GG+ GGG + G G +
Sbjct: 256 GDKGLQFCGQEGYFGGGGEGGGGGGGDGGGGGGDGHGSGGGG--------------GEGD 393
Query: 131 GGGGGG 136
GGGGGG
Sbjct: 394 GGGGGG 411
Score = 30.4 bits (67), Expect = 0.34
Identities = 19/45 (42%), Positives = 21/45 (46%), Gaps = 4/45 (8%)
Frame = +1
Query: 224 GGNG----RYDDGNGRFGGGNRRFGSGGGGHDGGKGTCFNCGKAG 264
GG G ++ G FGGG G GGGG DGG G G G
Sbjct: 247 GGGGDKGLQFCGQEGYFGGGGE--GGGGGGGDGGGGGGDGHGSGG 375
Score = 28.5 bits (62), Expect = 1.3
Identities = 15/32 (46%), Positives = 15/32 (46%)
Frame = +1
Query: 82 DNHGGGGGGRGFRGGERRNGGGGCYTCGDTGH 113
D HG GGGG GG GGGG GH
Sbjct: 355 DGHGSGGGGGEGDGG----GGGGSPQVSPQGH 438
>TC17113 weakly similar to UP|Q9LFU8 (Q9LFU8) Proline-rich protein, partial
(25%)
Length = 1077
Score = 42.4 bits (98), Expect = 9e-05
Identities = 33/90 (36%), Positives = 35/90 (38%)
Frame = -2
Query: 159 GGNNNNGGGGYGGGGTSCYRCGGVGHIARDCATPSSGGGGGGACYKCGEVGHIARDCSNE 218
GG N GGGG GGG GG G+ GGG G G +G
Sbjct: 785 GGKNGGGGGGGGGGVKPGITIGGKGNKGEGGC--FDGGGWNGLGINGGGLGGG*NGLGIS 612
Query: 219 GGRFDGGNGRYDDGNGRFGGGNRRFGSGGG 248
GG G NG G G GG N SGGG
Sbjct: 611 GGGLGG*NGLGKSGGG-LGGWNGSGKSGGG 525
Score = 42.0 bits (97), Expect = 1e-04
Identities = 38/112 (33%), Positives = 43/112 (37%), Gaps = 5/112 (4%)
Frame = -2
Query: 158 GGGNNN-----NGGGGYGGGGTSCYRCGGVGHIARDCATPSSGGGGGGACYKCGEVGHIA 212
GGGNN NGGGG GGG G + G G G C+ G +
Sbjct: 809 GGGNNGGKGGKNGGGGGGGG----------GGVKPGITIGGKGNKGEGGCFDGGGWNGLG 660
Query: 213 RDCSNEGGRFDGGNGRYDDGNGRFGGGNRRFGSGGGGHDGGKGTCFNCGKAG 264
N GG G NG G G GG G GGG G G+ GK+G
Sbjct: 659 ---INGGGLGGG*NGLGISGGGL--GG*NGLGKSGGGLGGWNGS----GKSG 531
Score = 41.2 bits (95), Expect = 2e-04
Identities = 58/199 (29%), Positives = 66/199 (33%), Gaps = 6/199 (3%)
Frame = -2
Query: 60 DNGKTKAVDVTGPKGEPLQVRQDNHGGGGGGRGFRGGERRNGGGGCYTCGDTGHIARDCD 119
D G+ V++ G G GG GG+G +NGGG
Sbjct: 851 DQGEEFFVEIGGLPG----------GGNNGGKG-----GKNGGG---------------- 765
Query: 120 RSDRNDRNDRSGGGGGGDRDRACYTCGSFEHFARDCMRGGGNNNNG--GGGYGGG----G 173
GGGGGG G C GGG N G GGG GGG G
Sbjct: 764 -----------GGGGGGGVKPGITIGGKGNKGEGGCFDGGGWNGLGINGGGLGGG*NGLG 618
Query: 174 TSCYRCGGVGHIARDCATPSSGGGGGGACYKCGEVGHIARDCSNEGGRFDGGNGRYDDGN 233
S GG + + SGGG GG N G+ GG G G
Sbjct: 617 ISGGGLGG*NGLGK------SGGGLGG---------------WNGSGKSGGGLG--*KGL 507
Query: 234 GRFGGGNRRFGSGGGGHDG 252
G GGG G GGG G
Sbjct: 506 GIRGGG*NGLGINGGG*KG 450
Database: LJGI
Posted date: Jul 30, 2004 11:16 AM
Number of letters in database: 14,692,800
Number of sequences in database: 28,460
Lambda K H
0.317 0.145 0.476
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 5,400,232
Number of Sequences: 28460
Number of extensions: 104015
Number of successful extensions: 3803
Number of sequences better than 10.0: 404
Number of HSP's better than 10.0 without gapping: 1545
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 2912
length of query: 275
length of database: 4,897,600
effective HSP length: 89
effective length of query: 186
effective length of database: 2,364,660
effective search space: 439826760
effective search space used: 439826760
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (22.0 bits)
S2: 54 (25.4 bits)
Medicago: description of AC121233.14


