

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC119416.7 - phase: 0 /pseudo
(447 letters)
Database: LJGI
28,460 sequences; 14,692,800 total letters
Searching..................................................done
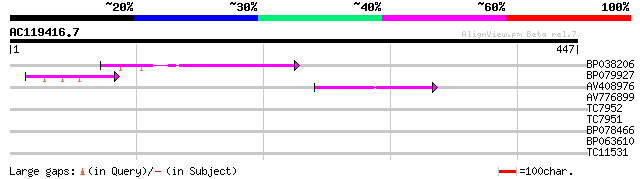
Score E
Sequences producing significant alignments: (bits) Value
BP038206 77 8e-15
BP079927 62 3e-10
AV408976 42 2e-04
AV776899 30 0.62
TC7952 similar to PIR|A45116|A45116 L-ascorbate peroxidase , cyt... 28 2.3
TC7951 similar to UP|Q43758 (Q43758) Ascorbate peroxidase , com... 28 2.3
BP078466 28 3.1
BP063610 27 6.8
TC11531 similar to AAQ56832 (AAQ56832) At2g34040, partial (32%) 27 6.8
>BP038206
Length = 563
Score = 76.6 bits (187), Expect = 8e-15
Identities = 51/169 (30%), Positives = 86/169 (50%), Gaps = 12/169 (7%)
Frame = -3
Query: 72 SSHGTTNLGKHLKVC--LKNPYRVVDKKQKTLA---------IGMGSEDDPNSVSFKLVD 120
S +GT +L H+ C +K R DK Q L +G G+ D NS
Sbjct: 540 SKNGTNHLRNHVAKCV*MKIQTRGHDKGQAFLMPKASQGKQELGAGAYDADNS------- 382
Query: 121 FSQEKTRLALAKMIIIDELPFKHVENEGFKMFMGEAQPKFKIPSRVSVARDCLRLYFDEK 180
K++LA A II+ + P V++ GF+ + QP F++P+R ++ ++ +++Y +K
Sbjct: 381 ----KSQLACA--IILHDYPLSIVDHIGFQRYSNSLQPLFQVPTRNTIKKEVIKIYEFQK 220
Query: 181 ETLKSVLSANNQMVSLTTDTWTS-IQNMNYMCVTAHYIDDEWVLKKKIL 228
+ +L ++ V++T+D WTS Q Y VT H ID W L+ ++L
Sbjct: 219 TCVMKMLDSHEGRVAITSDMWTSNNQKRGYTTVTTHCIDSTWTLQSRVL 73
>BP079927
Length = 438
Score = 61.6 bits (148), Expect = 3e-10
Identities = 33/88 (37%), Positives = 44/88 (49%), Gaps = 14/88 (15%)
Frame = +3
Query: 13 QGKDGEVTEAGAVT-----EVGAATAVSKQKKK---PQTCSKVWRHFNR------NKNLA 58
QG + E++ A EV A K+K P+T + +W+HF R A
Sbjct: 150 QGAETEISMADPAVLPPPPEVQGANPKKKRKPNASGPRTTTSIWKHFTRLPENEVQDPTA 329
Query: 59 VCKYCGKEYAANSSSHGTTNLGKHLKVC 86
C YCGK Y +S +HGTTNL HLK+C
Sbjct: 330 ACNYCGKRYLCDSKTHGTTNLNTHLKIC 413
>AV408976
Length = 340
Score = 42.0 bits (97), Expect = 2e-04
Identities = 30/97 (30%), Positives = 45/97 (45%)
Frame = +2
Query: 241 IGIALENCMKDWGIRSICCVTVDNASANNLAINYLNRGMRVWNGRTLFNGEYLHMRCCAH 300
I A+ C+ DW I N + + A+ L + V N L NG+ L C A
Sbjct: 50 ISHAVAVCLSDWNIEGRLFSITCNHALSESALGNLRSLLSVKNPLIL-NGQLLVGHCIAR 226
Query: 301 ILNLIVLDGLKDIDASVQRIRAACKFVKSSPSRLATF 337
L+ + D L ++ V++IR + KFVK+S S F
Sbjct: 227 TLSNVANDLLSAVEGIVKKIRDSVKFVKTSESHEEKF 337
>AV776899
Length = 396
Score = 30.4 bits (67), Expect = 0.62
Identities = 20/62 (32%), Positives = 28/62 (44%), Gaps = 7/62 (11%)
Frame = +2
Query: 34 VSKQKKKPQTC-SKVWRHF------NRNKNLAVCKYCGKEYAANSSSHGTTNLGKHLKVC 86
+ K K+P+ S VW+ F + A C C EY +GT+ L +H KVC
Sbjct: 212 LEKGTKRPKALTSDVWQKFTIIGRGDDGIQRAKCNGCRSEYKCGGD-YGTSTLRRHAKVC 388
Query: 87 LK 88
K
Sbjct: 389 TK 394
>TC7952 similar to PIR|A45116|A45116 L-ascorbate peroxidase , cytosolic
[validated] - garden pea {Pisum sativum;}
, partial (62%)
Length = 542
Score = 28.5 bits (62), Expect = 2.3
Identities = 18/57 (31%), Positives = 27/57 (46%)
Frame = -2
Query: 73 SHGTTNLGKHLKVCLKNPYRVVDKKQKTLAIGMGSEDDPNSVSFKLVDFSQEKTRLA 129
S GTT+LG H+K + P + K+ L SE P + LV + + RL+
Sbjct: 247 SEGTTSLGLHIKGPSRVPCQTEHKRSAFLVSDEASELSPGLLDSLLVISADGRVRLS 77
>TC7951 similar to UP|Q43758 (Q43758) Ascorbate peroxidase , complete
Length = 1022
Score = 28.5 bits (62), Expect = 2.3
Identities = 18/57 (31%), Positives = 27/57 (46%)
Frame = -2
Query: 73 SHGTTNLGKHLKVCLKNPYRVVDKKQKTLAIGMGSEDDPNSVSFKLVDFSQEKTRLA 129
S GTT+LG H+K + P + K+ L SE P + LV + + RL+
Sbjct: 241 SEGTTSLGLHIKGPSRVPCQTEHKRSAFLVSDEASELSPGLLDSLLVISADGRVRLS 71
>BP078466
Length = 530
Score = 28.1 bits (61), Expect = 3.1
Identities = 12/22 (54%), Positives = 17/22 (76%)
Frame = -1
Query: 128 LALAKMIIIDELPFKHVENEGF 149
+AL+ II+DE PF+ VE +GF
Sbjct: 290 VALSIFIILDEQPFRVVEGQGF 225
>BP063610
Length = 461
Score = 26.9 bits (58), Expect = 6.8
Identities = 9/24 (37%), Positives = 17/24 (70%)
Frame = -3
Query: 216 YIDDEWVLKKKILSFNLIADHKGE 239
Y+ D WVL + + + NL+ +++GE
Sbjct: 183 YLHDNWVLHRDLKTSNLLLNNRGE 112
>TC11531 similar to AAQ56832 (AAQ56832) At2g34040, partial (32%)
Length = 535
Score = 26.9 bits (58), Expect = 6.8
Identities = 11/32 (34%), Positives = 21/32 (65%)
Frame = +2
Query: 206 NMNYMCVTAHYIDDEWVLKKKILSFNLIADHK 237
N+ Y+C+T+ + + + + +LSF+LI HK
Sbjct: 149 NI*YLCLTSFQENGK*ICSEALLSFHLIQHHK 244
Database: LJGI
Posted date: Jul 30, 2004 11:16 AM
Number of letters in database: 14,692,800
Number of sequences in database: 28,460
Lambda K H
0.320 0.133 0.399
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 7,730,046
Number of Sequences: 28460
Number of extensions: 106533
Number of successful extensions: 502
Number of sequences better than 10.0: 18
Number of HSP's better than 10.0 without gapping: 495
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 500
length of query: 447
length of database: 4,897,600
effective HSP length: 93
effective length of query: 354
effective length of database: 2,250,820
effective search space: 796790280
effective search space used: 796790280
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 57 (26.6 bits)
Medicago: description of AC119416.7


