

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC149495.11 - phase: 0
(421 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
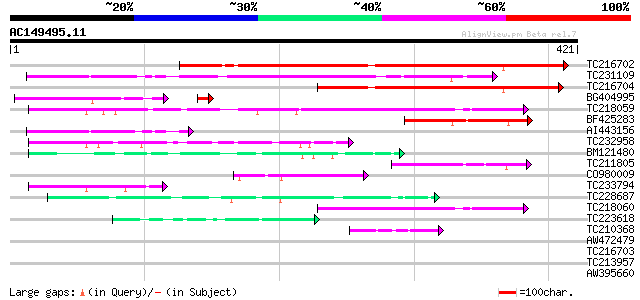
Score E
Sequences producing significant alignments: (bits) Value
TC216702 similar to UP|Q9LDP5 (Q9LDP5) FLORICAULA/LEAFY-like pro... 258 5e-69
TC231109 weakly similar to UP|Q9LUP7 (Q9LUP7) Gb|AAD25583.1, par... 239 1e-63
TC216704 similar to UP|Q7PIN1 (Q7PIN1) ENSANGP00000023661 (Fragm... 162 2e-40
BG404995 similar to GP|22831068|dbj OJ1131_E05.32 {Oryza sativa ... 92 1e-19
TC218059 weakly similar to UP|Q84KQ9 (Q84KQ9) F-box, partial (7%) 86 3e-17
BF425283 79 5e-15
AI443156 78 9e-15
TC232958 73 3e-13
BM121480 62 6e-10
TC211805 similar to UP|Q9K6E7 (Q9K6E7) BH3782 protein, partial (... 51 1e-06
CO980009 50 2e-06
TC233794 similar to GB|AAP37713.1|30725382|BT008354 At3g23880 {A... 49 4e-06
TC228687 similar to UP|Q9FGY4 (Q9FGY4) Gb|AAF30317.1, partial (79%) 49 6e-06
TC218060 47 2e-05
TC223618 42 4e-04
TC210368 42 7e-04
AW472479 weakly similar to PIR|T10479|T104 glycine-rich RNA-bind... 39 0.003
TC216703 39 0.004
TC213957 similar to GB|AAP37713.1|30725382|BT008354 At3g23880 {A... 38 0.007
AW395660 37 0.013
>TC216702 similar to UP|Q9LDP5 (Q9LDP5) FLORICAULA/LEAFY-like protein,
partial (5%)
Length = 1631
Score = 258 bits (658), Expect = 5e-69
Identities = 131/291 (45%), Positives = 193/291 (66%), Gaps = 2/291 (0%)
Frame = +3
Query: 127 NDATEITIWNTNTRKHWIIPFLPLPIPNIVESENIERGGVCIHGFGFDPLTGDYKLLRIS 186
N A +I WN + R+H I+P+LP+P ++ + VC GFGFD T DYKL+RIS
Sbjct: 15 NVADDIAFWNPSLRQHRILPYLPVPRRRHPDT-TLFAARVC--GFGFDHKTRDYKLVRIS 185
Query: 187 WLVSLQNSFYHSHARLFSSKTNSWKIIPIMPYTVYYAQAMGVFVENSIHWIMEKNLDGSD 246
+ V L + + + +L++ + N+WK +P +PY + A+ MGVFV NS+HW++ + L+
Sbjct: 186 YFVDLHDRSFDAQVKLYTLRANAWKTLPSLPYALCCARTMGVFVGNSLHWVVTRKLEPDQ 365
Query: 247 LCLIVAFNLTLEVFKEVPLPAVIRGEEVNNNQSFDLEVAVLGGCLSMIVNYQTTKIDVWV 306
LI+AF+LT ++F+E+PLP + + F++++A+LGG L M VN+ T+IDVWV
Sbjct: 366 PDLIIAFDLTHDIFRELPLP-----DTGGVDGGFEIDLALLGGSLCMTVNFHKTRIDVWV 530
Query: 307 MKEYGSRDSWCKIFTLVKSRFTLRLKSLRPLGYSSDGNKVLLQIDCKKLVWYDLKSEQV- 365
M+EY RDSWCK+FTL +SR LK +RPLGYSSDGNKVLL+ D K+L WYDL+ ++V
Sbjct: 531 MREYNRRDSWCKVFTLEESREMRSLKCVRPLGYSSDGNKVLLEHDRKRLFWYDLEKKEVA 710
Query: 366 -IYVEGIPNLDEAMICVESLVPPSFPVDNSSKKENRLSKSKRRYFFIDYLS 415
+ ++G+PNL+EAMIC+ +LVPP F +K L + R D+LS
Sbjct: 711 LVKIQGLPNLNEAMICLGTLVPPYFLPRQICRKPRALGCERPRKTRDDFLS 863
>TC231109 weakly similar to UP|Q9LUP7 (Q9LUP7) Gb|AAD25583.1, partial (8%)
Length = 1061
Score = 239 bits (611), Expect = 1e-63
Identities = 146/356 (41%), Positives = 212/356 (59%), Gaps = 6/356 (1%)
Frame = +3
Query: 13 SDLPPDILAEIFSCLPVKSLLRFRSTSKSLRSLIDSHKFINLHLKSSLNFKLILRHKTNL 72
++LP +++ EI S LPVKS++R RST K RS+IDS F+ HL S + LILRH+++L
Sbjct: 87 ANLPVEVVTEILSRLPVKSVIRLRSTCKWWRSIIDSRHFVLFHLNKS-HSSLILRHRSHL 263
Query: 73 YQLDFPNLTKSIIPLNHPFTTNIDPFTLNSIKALIGSCNGLLAIYNGPIAFTHPNDATEI 132
Y LD + ++ + L+HP NSIK L GS NGLL I N A +I
Sbjct: 264 YSLDLKSPEQNPVELSHPLMCYS-----NSIKVL-GSSNGLLCI---------SNVADDI 398
Query: 133 TIWNTNTRKHWIIPFLPLPIPNIVESENIERGGVCIHGFGFDPLTGDYKLLRISWLVSLQ 192
+WN RKH I+P P ++GFG + DYKLL I++ V LQ
Sbjct: 399 ALWNPFLRKHRILPADRFHRPQS------SLFAARVYGFGHHSPSNDYKLLSITYFVDLQ 560
Query: 193 NSFYHSHARLFSSKTNSWKIIPIMPYTVYYAQAMGVFVENSIHWIMEKNLDGSDLCLIVA 252
+ S +L++ K++SWK +P MPY + A+ MGVFV S+HW++ + L + LIV+
Sbjct: 561 KRTFDSQVQLYTLKSDSWKNLPSMPYALCCARTMGVFVSGSLHWLVTRKLQPHEPDLIVS 740
Query: 253 FNLTLEVFKEVPLPAVIRGEEVNNNQSFDLEVAVLGGCLSMIVNYQTTKIDVWVMKEYGS 312
F+LT E F EVPLP + G+ FD++VA+LGGCL +V ++ T DVWVM+ YGS
Sbjct: 741 FDLTRETFHEVPLPVTVNGD-------FDMQVALLGGCL-CVVEHRGTGFDVWVMRVYGS 896
Query: 313 RDSWCKIFTLVKSR------FTLRLKSLRPLGYSSDGNKVLLQIDCKKLVWYDLKS 362
R+SW K+FTL+++ + +LK +RPL + DG++VL + + KL WY+LK+
Sbjct: 897 RNSWEKLFTLLENNDHHEMMGSGKLKYVRPL--ALDGDRVLFEHNRIKLCWYNLKT 1058
>TC216704 similar to UP|Q7PIN1 (Q7PIN1) ENSANGP00000023661 (Fragment),
partial (5%)
Length = 1232
Score = 162 bits (411), Expect = 2e-40
Identities = 87/186 (46%), Positives = 121/186 (64%), Gaps = 3/186 (1%)
Frame = +2
Query: 229 FVENSIHWIMEKNLDGSDLCLIVAFNLTLEVFKEVPLPAVIRGEEVNNNQSFDLEVAVLG 288
FV NS+HW++ + L+ LIVAF+LT E+F E+PLP + F+++VA+LG
Sbjct: 2 FVGNSLHWVVTRKLEPDQPDLIVAFDLTHEIFTELPLP-----DTGGVGGGFEIDVALLG 166
Query: 289 GCLSMIVNYQTTKIDVWVMKEYGSRDSWCKIFTLVKSR-FTLRLKSLRPLGYSSDGNKVL 347
L M VN+ +K+DVWVM+EY DSWCK+FTL +S + LRPLGYSSDGNKVL
Sbjct: 167 DSLCMTVNFHNSKMDVWVMREYNRGDSWCKLFTLEESA*VEIVQXCLRPLGYSSDGNKVL 346
Query: 348 LQIDCKKLVWYDLKSEQV--IYVEGIPNLDEAMICVESLVPPSFPVDNSSKKENRLSKSK 405
L+ D K+L WYDL ++V + ++G+PNL+EAMIC+ +LV P F +K L +
Sbjct: 347 LEHDRKRLCWYDLGKKEVTLVRIQGLPNLNEAMICLGTLVTPYFLPRQICRKSPTLGCQR 526
Query: 406 RRYFFI 411
RR F+
Sbjct: 527 RRDDFL 544
>BG404995 similar to GP|22831068|dbj OJ1131_E05.32 {Oryza sativa (japonica
cultivar-group)}, partial (8%)
Length = 394
Score = 92.0 bits (227), Expect(2) = 1e-19
Identities = 59/119 (49%), Positives = 71/119 (59%), Gaps = 4/119 (3%)
Frame = +1
Query: 4 TLPSFLPVASDLPPDILAEIFSCLPVKSLLRFRSTSKSLRSLIDSHKFINLHLKSSL--- 60
T P L ++ LP ++L EI S LPV+SLLRFRSTSKS +SLIDS LHL SL
Sbjct: 7 TKPVLLTMSDHLPREVLTEILSRLPVRSLLRFRSTSKSWKSLIDSQHLNWLHLTRSLTLA 186
Query: 61 -NFKLILRHKTNLYQLDFPNLTKSIIPLNHPFTTNIDPFTLNSIKALIGSCNGLLAIYN 118
N LILR ++LYQ +FP L + LNHP + T L+GSCNGLL I N
Sbjct: 187 SNTSLILRVDSDLYQTNFPTLDPP-VSLNHPLMCYSNSIT------LLGSCNGLLCISN 342
Score = 22.3 bits (46), Expect(2) = 1e-19
Identities = 7/12 (58%), Positives = 11/12 (91%)
Frame = +3
Query: 140 RKHWIIPFLPLP 151
R+H I+P+LP+P
Sbjct: 345 RQHRILPYLPVP 380
>TC218059 weakly similar to UP|Q84KQ9 (Q84KQ9) F-box, partial (7%)
Length = 1185
Score = 85.9 bits (211), Expect = 3e-17
Identities = 93/395 (23%), Positives = 164/395 (40%), Gaps = 24/395 (6%)
Frame = +3
Query: 15 LPPDILAEIFSCLPVKSLLRFRSTSKSLRSLIDSHKFINLH------LKSSLNFKLILRH 68
LP ++++ + S LP K LL + KS LI F++ + L+S L++R
Sbjct: 66 LPGELVSNVLSRLPSKVLLLCKCVCKSWFDLITDPHFVSNYYVVYNSLQSQEEHLLVIRR 245
Query: 69 ------KTNLYQLDF----PNLTKSIIPLNHPFTTNIDPFTLNSIKALIGSCNGLLAIYN 118
KT + L + P S LN P+ N D I +G CNG+ +
Sbjct: 246 PFFSGLKTYISVLSWNTNDPKKHVSSDVLNPPYEYNSDHKYWTEI---LGPCNGIYFLEG 416
Query: 119 GPIAFTHPNDATEITIWNTNTRKHWIIPFLPLPIPNIVESENIERGGVCIHGFGFDPLTG 178
P +P+ + + + H+ P + GFGFDP T
Sbjct: 417 NPNVLMNPS----LGEFKALPKSHFTSPHGTYTFTDYA-------------GFGFDPKTN 545
Query: 179 DYKL--LRISWLVSL-QNSFYHSHARLFSSKTNSWK-----IIPIMPYTVYYAQAMGVFV 230
DYK+ L+ WL + + A L+S +NSW+ ++P+ P ++ + + +
Sbjct: 546 DYKVVVLKDLWLKETDEREIGYWSAELYSLNSNSWRKLDPSLLPL-PIEIWGSSRVFTYA 722
Query: 231 ENSIHWIMEKNLDGSDLCLIVAFNLTLEVFKEVPLPAVIRGEEVNNNQSFDLEVAVLGGC 290
N HW + +++AF++ E F+++ +P + + E + G
Sbjct: 723 NNCCHWWGFVEESDATQDVVLAFDMVKESFRKIRVPKIRDSSDEKFGTLVPFEESASIGF 902
Query: 291 LSMIVNYQTTKIDVWVMKEYGSRDSWCKIFTLVKSRFTLRLKSLRPLGYSSDGNKVLLQI 350
L V + DVWVMK+Y SW K +++ + +L +G+ N+ +
Sbjct: 903 LVYPVRGTEKRFDVWVMKDYWDEGSWVKQYSVGPVQVIYKL-----VGFYGT-NRFFWKD 1064
Query: 351 DCKKLVWYDLKSEQVIYVEGIPNLDEAMICVESLV 385
++LV YD ++ + + V G + A ESLV
Sbjct: 1065SNERLVLYDSENTRDLQVYGKHDSIRAAKYTESLV 1169
>BF425283
Length = 336
Score = 78.6 bits (192), Expect = 5e-15
Identities = 45/102 (44%), Positives = 64/102 (62%), Gaps = 7/102 (6%)
Frame = +1
Query: 294 IVNYQTTKIDVWVMKEYGSRDSWCKIFTLVKSRF----TLRLKSLRPLGYSSDGNKVLLQ 349
+V ++ T VWVM+ YGSRDSW K+F+L ++ + +LK +RPL DG++VL
Sbjct: 4 VVEHRGTGFHVWVMRVYGSRDSWEKLFSLTENHHHEMGSGKLKYVRPLAL-DDGDRVLFX 180
Query: 350 IDCKKLVWYDLKSEQVIYVE---GIPNLDEAMICVESLVPPS 388
+ KL WYDLK+ V V+ GI N E +CV+SLVPP+
Sbjct: 181 HNRXKLCWYDLKTGDVSCVKLPSGIGNTIERTVCVQSLVPPT 306
>AI443156
Length = 414
Score = 77.8 bits (190), Expect = 9e-15
Identities = 54/125 (43%), Positives = 70/125 (55%), Gaps = 1/125 (0%)
Frame = +3
Query: 13 SDLPPDILAEIFSCLPVKSLLRFRSTSKSLRSLIDSHKFINLHLKSSLNFKLILRHKTNL 72
++LP +++ EI S LPVKS++R RST K RS+IDS FI HL S + LILRH++ L
Sbjct: 84 ANLPVEVVTEILSRLPVKSVIRLRSTCKWWRSIIDSRHFILFHLNKS-HTSLILRHRSQL 260
Query: 73 YQLDFPN-LTKSIIPLNHPFTTNIDPFTLNSIKALIGSCNGLLAIYNGPIAFTHPNDATE 131
Y LD + L + L+HP NSIK L GS NGLL I N +
Sbjct: 261 YSLDLKSLLDPNPFELSHPLMC-----YSNSIKVL-GSSNGLLCI---------SNVXDD 395
Query: 132 ITIWN 136
I +WN
Sbjct: 396 IALWN 410
>TC232958
Length = 758
Score = 72.8 bits (177), Expect = 3e-13
Identities = 72/255 (28%), Positives = 116/255 (45%), Gaps = 14/255 (5%)
Frame = +1
Query: 15 LPPDILAEIFSCLPVKSLLRFRSTSKSLRSLIDSHKFINLH--LKSSLNFKL--ILRHKT 70
LP D++ EI LPVKSL+RF+S KS LI +F H L ++L ++ I
Sbjct: 61 LPQDLITEILLRLPVKSLVRFKSVCKSWLFLISDPRFAKSHFDLFAALADRILFIASSAP 240
Query: 71 NLYQLDFPNLTKSIIPLNHPFTTNID---PFTLNSIKALIGSCNGLLAIYNGPIAFTHPN 127
L +DF S+ + +D P +IGSC G + ++ +H
Sbjct: 241 ELRSIDF---NASLHDDSASVAVTVDLPAPKPYFHFVEIIGSCRGFILLH----CLSH-- 393
Query: 128 DATEITIWNTNTRKHWIIPFLPLPIPNIVESENIERGGVCIHGFGFDPLTGDYKLLRISW 187
+ +WN T H ++P P+ + + +C GFG+DP T D+ ++ +
Sbjct: 394 ----LCVWNPTTGVHKVVPLSPI----FFNKDAVFFTLLC--GFGYDPSTDDFLVVHACY 543
Query: 188 LVSLQNSFYHSHARLFSSKTNSWKIIP--IMPYTVY-----YAQAMGVFVENSIHWIMEK 240
Q + A +FS + N+WK I PYT + Y Q G F+ +IHW+ +
Sbjct: 544 NPKHQANC----AEIFSLRANAWKGIEGIHFPYTHFRYTNRYNQ-FGSFLNGAIHWLAFR 708
Query: 241 NLDGSDLCLIVAFNL 255
+ + +IVAF+L
Sbjct: 709 --INASINVIVAFDL 747
>BM121480
Length = 917
Score = 61.6 bits (148), Expect = 6e-10
Identities = 70/287 (24%), Positives = 115/287 (39%), Gaps = 8/287 (2%)
Frame = +3
Query: 15 LPPDILAEIFSCLPVKSLLRFRSTSKSLRSLIDSHKFINLHLKSSLNFKLILRHKTNLYQ 74
LP +++ EI LPVKSLLRF K +L +
Sbjct: 66 LPQELIREILLRLPVKSLLRF---------------------------KCVLFKTCSRDV 164
Query: 75 LDFPNLTKSIIPLNHPFTTNIDPFTLNSIKALIGSCNGLLAIYNGPIAFTHPNDATEITI 134
+ FP +PL +D F + ++GSC GL+ +Y +D+ + +
Sbjct: 165 VYFP------LPLPSIPCLRLDDFGIRP--KILGSCRGLVLLYY--------DDSANLIL 296
Query: 135 WNTNTRKHWIIPFLPLPIPNIVESENIERGGVCIHGFGFDPLTGDYKLLRISWLVSLQNS 194
WN + +H +P I + +GFG+D +Y L+ L+ L
Sbjct: 297 WNPSLGRHKXLPNYRDDITSFP------------YGFGYDESKDEYLLI----LIGLPKF 428
Query: 195 FYHSHARLFSSKTNSWKIIPIM--PYTVYYAQ----AMGVFVENSIHWIM--EKNLDGSD 246
+ A ++S KT SWK I+ P Y A+ G + ++HW + E D
Sbjct: 429 GPETGADIYSFKTESWKTDTIVYDPLXRYXAEDXIARAGSLLNGALHWFVFSESKXDH-- 602
Query: 247 LCLIVAFNLTLEVFKEVPLPAVIRGEEVNNNQSFDLEVAVLGGCLSM 293
+I+AF+L +PLP R V + + L+ ++G CL +
Sbjct: 603 --VIIAFDLVERTLSXIPLPLADR-STVQKDAVYGLK--IMGXCLXV 728
>TC211805 similar to UP|Q9K6E7 (Q9K6E7) BH3782 protein, partial (13%)
Length = 693
Score = 50.8 bits (120), Expect = 1e-06
Identities = 34/106 (32%), Positives = 54/106 (50%), Gaps = 2/106 (1%)
Frame = +1
Query: 284 VAVLGGCLSMIVNYQTTKIDVWVMKEYGSRDSWCKIFTLVKSRFTLRLKSLRPLGYSSDG 343
+ VL GCL M +Y+ T VW+MK+YG+R+SW K+ ++ P Y S+
Sbjct: 4 LGVLQGCLCMNYDYKKTHFVVWMMKDYGARESWVKLVSIPYVPNPENFSYSGPY-YISEN 180
Query: 344 NKVLLQIDCKKLVWYDLKSEQVIY--VEGIPNLDEAMICVESLVPP 387
+VLL + L+ Y+ + Y +E +A + VE+LV P
Sbjct: 181 GEVLLMFEF-DLILYNPRDNSFKYPKIESGKGWFDAEVYVETLVSP 315
>CO980009
Length = 736
Score = 50.1 bits (118), Expect = 2e-06
Identities = 35/107 (32%), Positives = 52/107 (47%), Gaps = 7/107 (6%)
Frame = -2
Query: 167 CIH----GFGFDPLTGDYKLLRISWLVSLQNSFYHSHA--RLFSSKTNSWKII-PIMPYT 219
C H GFD DYK++RIS +V SF S L+ T SW+I+ I P
Sbjct: 687 CYHHSYIALGFDSSNCDYKVIRISCIVD-DESFGLSAPLFELYPLATGSWRILDSISPVC 511
Query: 220 VYYAQAMGVFVENSIHWIMEKNLDGSDLCLIVAFNLTLEVFKEVPLP 266
A F + +HW+ ++ + G+ +++F L E+F EV LP
Sbjct: 510 YVAGDAPDGFEDGLVHWVAKRYVTGAWYYFVLSFRLEDEMFGEVMLP 370
>TC233794 similar to GB|AAP37713.1|30725382|BT008354 At3g23880 {Arabidopsis
thaliana;} , partial (9%)
Length = 449
Score = 48.9 bits (115), Expect = 4e-06
Identities = 38/114 (33%), Positives = 57/114 (49%), Gaps = 11/114 (9%)
Frame = +1
Query: 15 LPPDILAEIFSCLPVKSLLRFRSTSKSLRSLIDSHKFINLH--LKSSLNFKLILR-HKTN 71
LP +++ EI LPVKSLLRF+ KS SLI +F+ H L +S +LILR H
Sbjct: 109 LPQELIREILLRLPVKSLLRFKCVCKSFLSLISDPQFVISHYALAASPTHRLILRSHDFY 288
Query: 72 LYQLDFPNLTKSI--------IPLNHPFTTNIDPFTLNSIKALIGSCNGLLAIY 117
+ ++ K+ +PL +D F + ++GSC GL+ +Y
Sbjct: 289 AQSIATESVFKTCSRDVVYFPLPLPSIPCLRLDDFGIR--PKILGSCRGLVLLY 444
>TC228687 similar to UP|Q9FGY4 (Q9FGY4) Gb|AAF30317.1, partial (79%)
Length = 1228
Score = 48.5 bits (114), Expect = 6e-06
Identities = 67/302 (22%), Positives = 119/302 (39%), Gaps = 11/302 (3%)
Frame = +1
Query: 29 VKSLLRFRSTSKSLRSLIDSHKFINLHLKSSLNFKLILRHKTNLYQLDFPNLTKSIIPLN 88
VKSL RF++ K L F+ L+ + S +IL ++ + + T I N
Sbjct: 1 VKSLFRFKTVCKLWYRLSLDKYFVQLYNEVSRKNPMILVEISDSSE----SKTSLICVDN 168
Query: 89 HPFTTNIDPFTLNSIKALIGSCNGLLAIYNGPIAFTHPNDATEITIWNTNTRKHWIIPFL 148
+ LN + SCNGLL + P D + N TR++ ++P
Sbjct: 169 LRGVSEFSFIFLNDRVKVRASCNGLLCCSSIP-------DKGVFYVCNPVTREYRLLPKS 327
Query: 149 PLPIPNIVESENIER----GGVCIHGFGFDPLTGDYKLLRISWLVSLQNSFYHSH----- 199
++ R G + G D + ++ L F H
Sbjct: 328 --------RERHVTRFYPDGEATLVGLACDSAYRKFNVV----LAGYHRMFGHRPDGSFI 471
Query: 200 ARLFSSKTNSW-KIIPIMP-YTVYYAQAMGVFVENSIHWIMEKNLDGSDLCLIVAFNLTL 257
+F S+ N W K + + + + VFV N++HW+ + I+ +L+
Sbjct: 472 CLVFDSELNKWRKFVSFQDDHFTHMNKNQVVFVNNALHWLTASS------TYILVLDLSC 633
Query: 258 EVFKEVPLPAVIRGEEVNNNQSFDLEVAVLGGCLSMIVNYQTTKIDVWVMKEYGSRDSWC 317
EV++++ LP + N D + GCLS ++ +++WV+K+Y +D WC
Sbjct: 634 EVWRKMQLPYDLICGTGNRIYLLDFD-----GCLS-VIKISEAWMNIWVLKDYW-KDEWC 792
Query: 318 KI 319
+
Sbjct: 793 MV 798
>TC218060
Length = 901
Score = 47.0 bits (110), Expect = 2e-05
Identities = 39/157 (24%), Positives = 68/157 (42%)
Frame = +2
Query: 229 FVENSIHWIMEKNLDGSDLCLIVAFNLTLEVFKEVPLPAVIRGEEVNNNQSFDLEVAVLG 288
+ N HW G+ +++AF++ E F+++ +P V + E +
Sbjct: 11 YANNCCHWWGFVEDXGATQDIVLAFDMVKESFRKIRVPKVRDSSDEKFATLVPFEESASI 190
Query: 289 GCLSMIVNYQTTKIDVWVMKEYGSRDSWCKIFTLVKSRFTLRLKSLRPLGYSSDGNKVLL 348
G L V DVWVMK+Y SW K +++ + R+ +G+ N+ L
Sbjct: 191 GVLVYPVRGAEKSFDVWVMKDYWDEGSWVKQYSVGPVQVNHRI-----VGFYGT-NRFLW 352
Query: 349 QIDCKKLVWYDLKSEQVIYVEGIPNLDEAMICVESLV 385
+ ++LV YD + + + V G + A ESLV
Sbjct: 353 KDSNERLVLYDSEKTRDLQVYGKFDSIRAARYTESLV 463
>TC223618
Length = 444
Score = 42.4 bits (98), Expect = 4e-04
Identities = 41/154 (26%), Positives = 59/154 (37%)
Frame = +1
Query: 77 FPNLTKSIIPLNHPFTTNIDPFTLNSIKALIGSCNGLLAIYNGPIAFTHPNDATEITIWN 136
F NL+ LN+P ++GSCNGLL F D + +WN
Sbjct: 58 FNNLSTVCDELNYPVKNKFRH------DGIVGSCNGLL-------CFAIKGDC--VLLWN 192
Query: 137 TNTRKHWIIPFLPLPIPNIVESENIERGGVCIHGFGFDPLTGDYKLLRISWLVSLQNSFY 196
+ R P P+ N N G G G+D + DYK++ + F
Sbjct: 193 PSIRVSKKSP----PLGN-----NWRPGCFTAFGLGYDHVNEDYKVVAV--FCDPSEYFI 339
Query: 197 HSHARLFSSKTNSWKIIPIMPYTVYYAQAMGVFV 230
+++S TNSW+ I P+ Q G FV
Sbjct: 340 ECKVKVYSMATNSWRKIQDFPHGFSPFQNSGKFV 441
>TC210368
Length = 815
Score = 41.6 bits (96), Expect = 7e-04
Identities = 24/70 (34%), Positives = 35/70 (49%)
Frame = +1
Query: 253 FNLTLEVFKEVPLPAVIRGEEVNNNQSFDLEVAVLGGCLSMIVNYQTTKIDVWVMKEYGS 312
F+L E+PLP R V + + L + +GGCLS+ + ++WVMKEY
Sbjct: 1 FDLVERTLSEIPLPLADRST-VQKDAVYGLRI--MGGCLSVCCS-SCGMTEIWVMKEYKV 168
Query: 313 RDSWCKIFTL 322
R SW F +
Sbjct: 169 RSSWTMTFLI 198
>AW472479 weakly similar to PIR|T10479|T104 glycine-rich RNA-binding protein
GRP1 - Commerson's wild potato, partial (22%)
Length = 414
Score = 39.3 bits (90), Expect = 0.003
Identities = 45/153 (29%), Positives = 66/153 (42%), Gaps = 2/153 (1%)
Frame = +2
Query: 63 KLILRHKTNLYQLDFPNLTKSIIPLNHPFTTNIDPFTLNSIKALIGSCNGLLAIYNGPIA 122
KL L H+ + + P+LT S L P +++ ++ CNGL+ I G
Sbjct: 20 KLFLPHRRHHHD---PSLTLSYTLLRLPSFPDLE-------FPVLSFCNGLICIAYG--- 160
Query: 123 FTHPNDATEITIWNTNTRKHWIIPFLPLPIPNIVESENIERGGVCIHGFGFDPLTGDYKL 182
I I N + R++ +P P P S CI GFD DYK+
Sbjct: 161 ----ERCLPIIICNPSIRRYVWLP-TPHDYPGYYNS--------CI-ALGFDSTNCDYKV 298
Query: 183 LRISWLVSLQNSFYHSH--ARLFSSKTNSWKII 213
+RIS +V SF S L+S K+ SW+I+
Sbjct: 299 IRISCIVD-DESFGLSAPLVELYSLKSGSWRIL 394
>TC216703
Length = 776
Score = 38.9 bits (89), Expect = 0.004
Identities = 19/51 (37%), Positives = 30/51 (58%)
Frame = +2
Query: 361 KSEQVIYVEGIPNLDEAMICVESLVPPSFPVDNSSKKENRLSKSKRRYFFI 411
K ++ ++G+PNL+EAMIC+ +LV P F +K L +RR F+
Sbjct: 2 KEVTLVRIQGLPNLNEAMICLGTLVTPYFLPRQICRKSPTLGCQRRRDDFL 154
>TC213957 similar to GB|AAP37713.1|30725382|BT008354 At3g23880 {Arabidopsis
thaliana;} , partial (9%)
Length = 696
Score = 38.1 bits (87), Expect = 0.007
Identities = 21/52 (40%), Positives = 31/52 (59%)
Frame = +3
Query: 15 LPPDILAEIFSCLPVKSLLRFRSTSKSLRSLIDSHKFINLHLKSSLNFKLIL 66
LP +++ EI PV+S+LRF+ KS SLI +F + L +S +LIL
Sbjct: 33 LPLELIREILLRSPVRSVLRFKCVCKSWLSLISDPQFTHFDLAASPTHRLIL 188
>AW395660
Length = 381
Score = 37.4 bits (85), Expect = 0.013
Identities = 19/32 (59%), Positives = 23/32 (71%)
Frame = +3
Query: 15 LPPDILAEIFSCLPVKSLLRFRSTSKSLRSLI 46
LP +++ EI S LPVKSLL+FR KS SLI
Sbjct: 264 LPDELVVEILSRLPVKSLLQFRCVCKSWMSLI 359
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.323 0.140 0.429
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 22,121,973
Number of Sequences: 63676
Number of extensions: 353587
Number of successful extensions: 1886
Number of sequences better than 10.0: 52
Number of HSP's better than 10.0 without gapping: 1853
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1865
length of query: 421
length of database: 12,639,632
effective HSP length: 100
effective length of query: 321
effective length of database: 6,272,032
effective search space: 2013322272
effective search space used: 2013322272
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.5 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 60 (27.7 bits)
Medicago: description of AC149495.11


