

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC149494.4 - phase: 0
(291 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
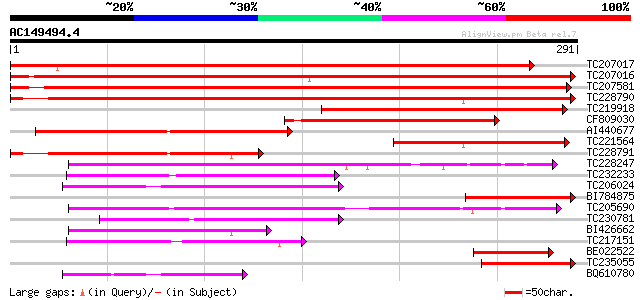
Score E
Sequences producing significant alignments: (bits) Value
TC207017 447 e-126
TC207016 437 e-123
TC207581 363 e-101
TC228790 311 2e-85
TC219918 182 2e-46
CF809030 149 1e-36
AI440677 130 9e-31
TC221564 115 2e-26
TC228791 107 8e-24
TC228247 weakly similar to UP|Q8LBR0 (Q8LBR0) Phloem-specific le... 103 7e-23
TC232233 weakly similar to GB|AAT41865.1|48310676|BT014882 At1g8... 98 4e-21
TC206024 weakly similar to UP|Q9FLU7 (Q9FLU7) Phloem-specific le... 97 1e-20
BI784875 93 2e-19
TC205690 weakly similar to UP|Q6K3E9 (Q6K3E9) F-box family prote... 92 2e-19
TC230781 weakly similar to UP|Q6K3E9 (Q6K3E9) F-box family prote... 74 6e-14
BI426662 66 2e-11
TC217151 similar to UP|Q6NPT8 (Q6NPT8) At2g02230, partial (6%) 66 2e-11
BE022522 59 3e-09
TC235055 57 8e-09
BQ610780 52 3e-07
>TC207017
Length = 905
Score = 447 bits (1150), Expect = e-126
Identities = 215/271 (79%), Positives = 237/271 (87%), Gaps = 2/271 (0%)
Frame = +3
Query: 1 MGASISSTASIREKNNEESNSSS--SKTGLGDIPESCISSILMHLDPHDICKLARVNRAF 58
MGA +SST + +SSS S+ GLGDIPESCISS+ M+LDP DICKLARVNRAF
Sbjct: 93 MGAGVSSTNKAEKDGGGSISSSSRDSRPGLGDIPESCISSLFMNLDPPDICKLARVNRAF 272
Query: 59 HRASSADFVWESKLPSCYKFLANKFLGEENLSTMSKKDVYTKLCQPNRFDGGTKEVLLDK 118
HRASSADFVWESKLP YKFLANK LGEEN++TM+KK++Y KLC PNRFDGGTKEV LDK
Sbjct: 273 HRASSADFVWESKLPPSYKFLANKVLGEENIATMTKKEIYAKLCLPNRFDGGTKEVWLDK 452
Query: 119 YSGQVCLFMSSKSLKITGIDDRRYWIYIPTEESRFKNVAYLQQMWWVEVIGELEFVFPVG 178
SGQVCLFMSSKSLKITGIDDRRYW YIPTEESRF++VAYLQQMWWVEV+GELEF FPVG
Sbjct: 453 CSGQVCLFMSSKSLKITGIDDRRYWNYIPTEESRFQSVAYLQQMWWVEVVGELEFEFPVG 632
Query: 179 SYSITFRLQLGKSSKRLGRRVCNDDQVHGWDIKPVRFQLSTSDGQRSISECYLHEPGQWA 238
SYS+ FRLQLGK+SKRLGRRVCN DQVHGWDIKP+RFQLSTSDGQ S+SECYL PG+W
Sbjct: 633 SYSLIFRLQLGKASKRLGRRVCNVDQVHGWDIKPIRFQLSTSDGQSSLSECYLRGPGEWV 812
Query: 239 YYHVGDFMVTKPNKPIKIKFSLAQIDCTHTK 269
YY+VGDF+V KP +PI IKFSLAQIDCTHTK
Sbjct: 813 YYNVGDFVVEKPKEPINIKFSLAQIDCTHTK 905
>TC207016
Length = 1310
Score = 437 bits (1124), Expect = e-123
Identities = 211/293 (72%), Positives = 244/293 (83%), Gaps = 3/293 (1%)
Frame = +1
Query: 1 MGASISSTASIREKNNEESNSSSSKTGLGDIPESCISSILMHLDPHDICKLARVNRAFHR 60
MGA +S+TA+ E ++ + S SS+T L DIPE+CISS++M+ DP +IC LARVN+ FHR
Sbjct: 232 MGAGVSTTAT--ENDDGGTFSLSSETSLDDIPENCISSMMMNFDPQEICSLARVNKTFHR 405
Query: 61 ASSADFVWESKLPSCYKFLANKFLGEENLSTMSKKDVYTKLCQPNRFDGGTKEVLLDKYS 120
ASSA+FVWESKLP YKFL NK LGE+NL +M+KK++Y KLCQPN FDGGTKEV LD+ S
Sbjct: 406 ASSANFVWESKLPQNYKFLLNKVLGEQNLGSMTKKEIYAKLCQPNFFDGGTKEVWLDRSS 585
Query: 121 GQVCLFMSSKSLKITGIDDRRYWIYIPTEESR---FKNVAYLQQMWWVEVIGELEFVFPV 177
GQVC+F+SSKS KITGIDDRRYW YIPTEESR FK+VAYLQQMWWVEVIGELEF FP
Sbjct: 586 GQVCMFISSKSFKITGIDDRRYWNYIPTEESR*ERFKSVAYLQQMWWVEVIGELEFEFPK 765
Query: 178 GSYSITFRLQLGKSSKRLGRRVCNDDQVHGWDIKPVRFQLSTSDGQRSISECYLHEPGQW 237
G+YS+ F+LQLGK SKRLGRRVCN DQVHGWDIKPVRFQLSTSDGQRS+S+CYL +W
Sbjct: 766 GNYSVFFKLQLGKPSKRLGRRVCNLDQVHGWDIKPVRFQLSTSDGQRSLSQCYLRGSREW 945
Query: 238 AYYHVGDFMVTKPNKPIKIKFSLAQIDCTHTKGGLCIDSAIIRPTEFRERLKK 290
AYYHVGDF + KPN P I FSLAQIDCTHTKGGLCID +I P EF + LK+
Sbjct: 946 AYYHVGDFAIDKPNGPTNINFSLAQIDCTHTKGGLCIDGVVICPKEFTQGLKQ 1104
>TC207581
Length = 1562
Score = 363 bits (931), Expect = e-101
Identities = 169/288 (58%), Positives = 217/288 (74%)
Frame = +2
Query: 1 MGASISSTASIREKNNEESNSSSSKTGLGDIPESCISSILMHLDPHDICKLARVNRAFHR 60
MGAS SS + + + + LGDIPESC++ +LM+LDP DICKLAR+NRAF
Sbjct: 395 MGASFSSCVC-------DGDGTPLRPRLGDIPESCVALVLMYLDPPDICKLARLNRAFRD 553
Query: 61 ASSADFVWESKLPSCYKFLANKFLGEENLSTMSKKDVYTKLCQPNRFDGGTKEVLLDKYS 120
ASSADF+WESKLP YKF+ K L + ++ + K+D+Y +LC+PN FD GTKE+ LDK +
Sbjct: 554 ASSADFIWESKLPLNYKFIVEKALKDVSVEQLGKRDIYARLCRPNSFDNGTKEIWLDKRT 733
Query: 121 GQVCLFMSSKSLKITGIDDRRYWIYIPTEESRFKNVAYLQQMWWVEVIGELEFVFPVGSY 180
G VCL +SS++L+ITGIDDRRYW I TEESRF VAYLQQ+WW+EV +++F FP G Y
Sbjct: 734 GGVCLAISSQALRITGIDDRRYWSRISTEESRFHTVAYLQQIWWLEVEDDVDFQFPPGKY 913
Query: 181 SITFRLQLGKSSKRLGRRVCNDDQVHGWDIKPVRFQLSTSDGQRSISECYLHEPGQWAYY 240
S+ FRLQLG+SSKRLGRRVC D +HGWDIKPV+FQL+TSDGQR++S+ +L PG W Y
Sbjct: 914 SVFFRLQLGRSSKRLGRRVCKTDDIHGWDIKPVKFQLTTSDGQRAVSQSHLDNPGHWVLY 1093
Query: 241 HVGDFMVTKPNKPIKIKFSLAQIDCTHTKGGLCIDSAIIRPTEFRERL 288
H G+F+ PN +KIKFSL QIDCTHTKGGLC+DS I ++ ++ +
Sbjct: 1094HAGNFVSKSPNDLMKIKFSLTQIDCTHTKGGLCVDSVFICNSDLKKEV 1237
>TC228790
Length = 1215
Score = 311 bits (798), Expect = 2e-85
Identities = 155/302 (51%), Positives = 208/302 (68%), Gaps = 12/302 (3%)
Frame = +2
Query: 1 MGASISSTASIREKNNEESNSSSSKTGLGDIPESCISSILMHLDPHDICKLARVNRAFHR 60
MGAS+SS N SS GLGDIPESC++ + +HL P +IC LAR+NRAF
Sbjct: 131 MGASLSS------------NGSSIGPGLGDIPESCVACVFLHLTPPEICNLARLNRAFRG 274
Query: 61 ASSADFVWESKLPSCYKFLANKFLG-EENLSTMSKKDVYTKLCQPNRFDGGTKEVLLDKY 119
A+S+D VWE+KLP Y+ L + E + ++SKKD++ L +P FD G KEV LD+
Sbjct: 275 AASSDSVWEAKLPRNYQDLLDLVPPPERHHRSLSKKDIFALLSRPLPFDHGHKEVWLDRV 454
Query: 120 SGQVCLFMSSKSLKITGIDDRRYWIYIPTEESRFKNVAYLQQMWWVEVIGELEFVFPVGS 179
+G+VC+ +S+K++ ITGIDDRRYW +IPTEESRF VAYLQQ+WW EV GE+ F FP
Sbjct: 455 TGKVCMSISAKAMVITGIDDRRYWNWIPTEESRFHTVAYLQQIWWFEVDGEVTFPFPADI 634
Query: 180 YSITFRLQLGKSSKRLGRRVCNDDQVHGWDIKPVRFQLSTSDGQRSISECYL-------- 231
Y+++FRL LG+ SKRLGRRVCN + HGWDIKPV+F+ STSDGQ++ EC L
Sbjct: 635 YTLSFRLHLGRFSKRLGRRVCNYEHTHGWDIKPVKFEFSTSDGQQASCECCLDETEINDT 814
Query: 232 ---HEPGQWAYYHVGDFMVTKPNKPIKIKFSLAQIDCTHTKGGLCIDSAIIRPTEFRERL 288
H+ G W Y VG+F+V+ +++FS+ QIDCTH+KGGLC+D+ I P++ RER
Sbjct: 815 YGNHKRGYWVDYKVGEFIVSGSEPTTQVRFSMKQIDCTHSKGGLCVDAVFIVPSDLRERK 994
Query: 289 KK 290
++
Sbjct: 995 RR 1000
>TC219918
Length = 661
Score = 182 bits (461), Expect = 2e-46
Identities = 77/126 (61%), Positives = 98/126 (77%)
Frame = +2
Query: 161 QMWWVEVIGELEFVFPVGSYSITFRLQLGKSSKRLGRRVCNDDQVHGWDIKPVRFQLSTS 220
Q+WW +V GE+EF FP G YS+ FR+ LG++ KR GRRVCN + VHGWD KPVRFQL TS
Sbjct: 2 QIWWFQVDGEVEFPFPAGKYSVFFRIHLGRAGKRCGRRVCNTEHVHGWDKKPVRFQLWTS 181
Query: 221 DGQRSISECYLHEPGQWAYYHVGDFMVTKPNKPIKIKFSLAQIDCTHTKGGLCIDSAIIR 280
DGQ S+C+L+ PG+W +YH GDF+V N K+KFS+ QIDCTHTKGGLC+DS ++
Sbjct: 182 DGQYVASQCFLNGPGKWIFYHAGDFVVEDGNASTKVKFSMTQIDCTHTKGGLCLDSVLVY 361
Query: 281 PTEFRE 286
P+EFR+
Sbjct: 362 PSEFRK 379
>CF809030
Length = 762
Score = 149 bits (376), Expect = 1e-36
Identities = 66/110 (60%), Positives = 84/110 (76%)
Frame = +1
Query: 142 YWIYIPTEESRFKNVAYLQQMWWVEVIGELEFVFPVGSYSITFRLQLGKSSKRLGRRVCN 201
Y I++ +RF VAYLQQ+WW+EV G+++F FP GSY++ FRLQLG+SSKRLGRRVC
Sbjct: 433 YTIFV---HNRFHTVAYLQQIWWLEVEGDVDFQFPPGSYNVFFRLQLGRSSKRLGRRVCK 603
Query: 202 DDQVHGWDIKPVRFQLSTSDGQRSISECYLHEPGQWAYYHVGDFMVTKPN 251
D VHGWDIKPV+FQL+TSDGQ ++S+ +L PG W YH G F+ PN
Sbjct: 604 TDDVHGWDIKPVKFQLTTSDGQHAVSQSHLDNPGNWILYHAGXFVSKNPN 753
>AI440677
Length = 421
Score = 130 bits (326), Expect = 9e-31
Identities = 62/132 (46%), Positives = 92/132 (68%)
Frame = +1
Query: 14 KNNEESNSSSSKTGLGDIPESCISSILMHLDPHDICKLARVNRAFHRASSADFVWESKLP 73
++N SN S++ GLGDIPE+C++ + +HL P +IC LAR+NRAF A+SAD VW++KLP
Sbjct: 4 RSNLGSNGSAAAPGLGDIPENCVARVFLHLTPPEICNLARLNRAFRGAASADSVWQTKLP 183
Query: 74 SCYKFLANKFLGEENLSTMSKKDVYTKLCQPNRFDGGTKEVLLDKYSGQVCLFMSSKSLK 133
Y+ L + + E +SKKD++ L + FD G K V LD+ + +VC+ +S+K++
Sbjct: 184 RNYQDLLD-LMPPERHRNLSKKDIFALLSRAVPFDDGNKGVWLDRVTRRVCMSISAKAMS 360
Query: 134 ITGIDDRRYWIY 145
ITGIDDRRYW +
Sbjct: 361 ITGIDDRRYWTW 396
>TC221564
Length = 697
Score = 115 bits (289), Expect = 2e-26
Identities = 53/101 (52%), Positives = 68/101 (66%), Gaps = 11/101 (10%)
Frame = +3
Query: 198 RVCNDDQVHGWDIKPVRFQLSTSDGQRSISECYL-----------HEPGQWAYYHVGDFM 246
RVC+ + HGWDIKPVRF+LST DGQ++ SECYL H+ G W Y VG+F+
Sbjct: 21 RVCSYEHTHGWDIKPVRFELSTMDGQQASSECYLDETEPDDLHGNHKRGHWVDYKVGEFI 200
Query: 247 VTKPNKPIKIKFSLAQIDCTHTKGGLCIDSAIIRPTEFRER 287
V+ K++FS+ QIDCTH+KGGLC+DS I P + RER
Sbjct: 201 VSGSEPTTKVRFSMKQIDCTHSKGGLCVDSVFIIPRDLRER 323
>TC228791
Length = 584
Score = 107 bits (266), Expect = 8e-24
Identities = 61/136 (44%), Positives = 85/136 (61%), Gaps = 6/136 (4%)
Frame = +1
Query: 1 MGASISSTASIREKNNEESNSSSSKTGLGDIPESCISSILMHLDPHDICKLARVNRAFHR 60
MGAS+SS N SS GLGDIPESC++ + +HL P +IC LAR+NRAF
Sbjct: 214 MGASLSS------------NGSSIGPGLGDIPESCVACVFLHLTPPEICNLARLNRAFRG 357
Query: 61 ASSADFVWESKLPSCYKFLANKFLGEENLSTMSKKDVYTKLCQPNRFDGGTK------EV 114
A+S+D VWE+KLP Y+ L + + E ++SKKD++ L +P FD G K EV
Sbjct: 358 AASSDSVWEAKLPRNYQDLLD-LVPPERHRSLSKKDIFALLSRPLPFDHGHKVGAALQEV 534
Query: 115 LLDKYSGQVCLFMSSK 130
LD+ +G+VC+ +S+K
Sbjct: 535 WLDRVTGKVCMSISAK 582
>TC228247 weakly similar to UP|Q8LBR0 (Q8LBR0) Phloem-specific lectin
PP2-like protein, partial (46%)
Length = 1124
Score = 103 bits (258), Expect = 7e-23
Identities = 76/265 (28%), Positives = 127/265 (47%), Gaps = 14/265 (5%)
Frame = +3
Query: 31 IPESCISSILMHLDPHDICKLARVNRAFHRASSADFVWESKLPSCYKFLANKFLGEENL- 89
+PE C+S IL + P D C+ + V+ ++ +D +W + PS Y + ++ L +L
Sbjct: 81 LPEDCVSKILSYTSPPDACRFSMVSSTLRSSADSDLLWRTFFPSDYSDIVSRALNPLSLN 260
Query: 90 STMSKKDVYTKLCQPNRFDGGTKEVLLDKYSGQVCLFMSSKSLKITGIDDRRYWIYIPTE 149
S+ S K ++ LC P DGG LDK SG+ +S++ L IT +D YW + P
Sbjct: 261 SSSSYKHLFYALCHPLLLDGGNMSFKLDKSSGKKSYILSARQLSITWSNDPLYWSWRPVP 440
Query: 150 ESRFKNVAYLQQMWWVEVIGEL--EFVFPVGSYSI-----TFRLQLGKSSKRLGRRVCND 202
ESRFK VA L+ + W+E+ G++ + P SY + T + G S + D
Sbjct: 441 ESRFKEVAELRTVSWLEIQGKIGTRILTPNTSYVVYLIMKTSHREYGLDSVACEVSIAVD 620
Query: 203 DQVHGWDIKPVRFQLSTSD------GQRSISECYLHEPGQWAYYHVGDFMVTKPNKPIKI 256
++V + R L ++ + SI E G W +G+F + ++ ++
Sbjct: 621 NKVK----QSGRVYLCQNEKDENNLKKESIGIPMRREDG-WMEIEMGEFFCGEADE--EV 779
Query: 257 KFSLAQIDCTHTKGGLCIDSAIIRP 281
SL ++ KGGL ++ IRP
Sbjct: 780 LMSLMEVG-YQLKGGLIVEGVEIRP 851
>TC232233 weakly similar to GB|AAT41865.1|48310676|BT014882 At1g80110
{Arabidopsis thaliana;} , partial (47%)
Length = 687
Score = 98.2 bits (243), Expect = 4e-21
Identities = 51/141 (36%), Positives = 81/141 (57%), Gaps = 1/141 (0%)
Frame = +1
Query: 30 DIPESCISSILMHLDPHDICKLARVNRAFHRASSADFVWESKLPSCYKFLANKFLGEENL 89
++PE CI++IL P D+C+L+ ++ F A+ +D VW LPS + + ++ +L
Sbjct: 22 NLPEGCIANILSFTSPRDVCRLSLLSSTFRSAAQSDAVWNKFLPSDFHTILSQ---SSSL 192
Query: 90 STMSKKDVYTKLCQ-PNRFDGGTKEVLLDKYSGQVCLFMSSKSLKITGIDDRRYWIYIPT 148
S SKKD++ LCQ P D G K LDK G+ C +S+++L I D RYW +
Sbjct: 193 SLPSKKDLFLYLCQKPLLIDDGKKSYQLDKVYGKKCYMLSARNLFIVWGDTPRYWRWTSL 372
Query: 149 EESRFKNVAYLQQMWWVEVIG 169
++RF VA L+ + W+E+ G
Sbjct: 373 PDARFSEVAELRSVCWLEIRG 435
>TC206024 weakly similar to UP|Q9FLU7 (Q9FLU7) Phloem-specific lectin-like
protein, partial (47%)
Length = 954
Score = 96.7 bits (239), Expect = 1e-20
Identities = 53/145 (36%), Positives = 82/145 (56%), Gaps = 1/145 (0%)
Frame = +3
Query: 28 LGDIPESCISSILMHLDPHDICKLARVNRAFHRASSADFVWESKLPSCYKFLANKFLGEE 87
L D+PE CI+ IL + P D+C+L+ V++AF A+ +D VW+ FL + F
Sbjct: 27 LQDLPEGCIAKILSYTTPVDVCRLSLVSKAFRSAAESDTVWDC-------FLLSDFTSII 185
Query: 88 NLSTMSKKDVYTKLCQ-PNRFDGGTKEVLLDKYSGQVCLFMSSKSLKITGIDDRRYWIYI 146
+S+ SKKD+Y L P G K V LDK +G+ C +S+++L I D ++W +
Sbjct: 186 PISSTSKKDLYFTLSDHPTIIHQGRKSVQLDKRTGKKCCMLSARNLTIIWGDTVQHWEWT 365
Query: 147 PTEESRFKNVAYLQQMWWVEVIGEL 171
ESRF+ VA LQ + W ++ G +
Sbjct: 366 SLPESRFQEVAMLQAVCWFDISGSI 440
>BI784875
Length = 322
Score = 92.8 bits (229), Expect = 2e-19
Identities = 40/56 (71%), Positives = 45/56 (79%)
Frame = +1
Query: 235 GQWAYYHVGDFMVTKPNKPIKIKFSLAQIDCTHTKGGLCIDSAIIRPTEFRERLKK 290
G+WA+YHVGDF + KPN P I FSLAQIDCTHTKGGLCID +I P EF ERLK+
Sbjct: 1 GEWAHYHVGDFAIDKPNGPTNINFSLAQIDCTHTKGGLCIDGVVICPKEFTERLKQ 168
>TC205690 weakly similar to UP|Q6K3E9 (Q6K3E9) F-box family protein-like,
partial (33%)
Length = 1360
Score = 92.4 bits (228), Expect = 2e-19
Identities = 71/273 (26%), Positives = 122/273 (44%), Gaps = 20/273 (7%)
Frame = +2
Query: 31 IPESCISSILMHLDPHDICKLARVNRAFHRASSADFVWESKLPSCYKFLANKFLGEENLS 90
+PE CI+SIL P D+C+ + V++ F A+ +D VW+ LPS Y + ++ L+
Sbjct: 215 LPEGCIASILSRTTPADVCRFSVVSKIFRSAAESDAVWKRFLPSDYHSIISQ--SPSPLN 388
Query: 91 TMSKKDVYTKLC-QPNRFDGGTKEVLLDKYSGQVCLFMSSKSLKITGIDDRRYWIYIPTE 149
SKK++Y L +P D G K L+K SG+ C +++++L I D +YW +
Sbjct: 389 YPSKKELYLALSDRPIIIDQGKKSFQLEKKSGKKCYMLAARALSIIWGDTEQYWNWTTDT 568
Query: 150 ESRFKNVAYLQQMWWVEVIGELEFVFPVGSYSITFRLQLGKSSKRLGRRVCNDDQVHGWD 209
SRF VA L+ + W+E+ G L L L +++ V G+
Sbjct: 569 NSRFPEVAELRDVCWLEIRGVLN------------TLVLSPNTQYAAYLVFKMIDARGFH 712
Query: 210 IKPVRFQLSTSDGQRSISECYLHEPGQ-------------------WAYYHVGDFMVTKP 250
+PV ++ G S L +P + W +G+F T
Sbjct: 713 NRPVELSVNVFGGHGSTKIVCL-DPNEELPHRRVEGLQRPNARSDGWLEIEMGEFFNTGL 889
Query: 251 NKPIKIKFSLAQIDCTHTKGGLCIDSAIIRPTE 283
+ +++ S+ + + K GL I+ ++P E
Sbjct: 890 DD--EVQMSVVETKGGNWKSGLFIEGIEVKPKE 982
>TC230781 weakly similar to UP|Q6K3E9 (Q6K3E9) F-box family protein-like,
partial (29%)
Length = 971
Score = 74.3 bits (181), Expect = 6e-14
Identities = 41/126 (32%), Positives = 67/126 (52%), Gaps = 1/126 (0%)
Frame = +2
Query: 47 DICKLARVNRAFHRASSADFVWESKLPSCYKFLANKFLGEENLSTMSKKDVYTKLC-QPN 105
D+C+L+ V++AFH A+ A+ VW+ LPS + + ++ KD+Y L +P
Sbjct: 14 DVCRLSLVSKAFHSAAEANTVWDCFLPSDLSSIISPSSSVPPFRSI--KDLYLYLSDRPT 187
Query: 106 RFDGGTKEVLLDKYSGQVCLFMSSKSLKITGIDDRRYWIYIPTEESRFKNVAYLQQMWWV 165
D G K L+K +G C +S++ L I D YW + ESRF+ VA L+ + W
Sbjct: 188 IIDQGIKSFQLEKRTGNKCYMLSARDLSIIWGDTTHYWEWTTLPESRFEEVARLRAVCWF 367
Query: 166 EVIGEL 171
++ G +
Sbjct: 368 DITGRM 385
>BI426662
Length = 421
Score = 65.9 bits (159), Expect = 2e-11
Identities = 43/133 (32%), Positives = 62/133 (46%), Gaps = 29/133 (21%)
Frame = +3
Query: 31 IPESCISSILMHLDPHDICKLARVNRAFHRASSADFVWESKLPSCYKFLANKFLGEENLS 90
+PE CI++I+ P D C L+ V+ +F AS DFVWE LPS Y+ + ++ L+
Sbjct: 21 LPEGCIANIVSFTTPPDACVLSLVSSSFRSASVTDFVWERFLPSDYQAIISQSSKPSTLT 200
Query: 91 T-MSKKDVYTKLC-QPNRFDGGTK---------------------------EVLLDKYSG 121
SKKD+Y LC P D G K LDK +G
Sbjct: 201 NYSSKKDLYLHLCHNPLLIDAGKKVLQIKSFFLSIIIHVYLSLIGKLINHQSFALDKLNG 380
Query: 122 QVCLFMSSKSLKI 134
++C +S++SL I
Sbjct: 381 KICYMLSARSLSI 419
>TC217151 similar to UP|Q6NPT8 (Q6NPT8) At2g02230, partial (6%)
Length = 514
Score = 65.9 bits (159), Expect = 2e-11
Identities = 41/129 (31%), Positives = 69/129 (52%), Gaps = 6/129 (4%)
Frame = +3
Query: 30 DIPESCISSILMHL-DPHDICKLARVNRAFHRASSADFVWESKLPSCYKFLANKFLGEEN 88
D+PE C++ IL ++ P DI +L+ V++AF+ A+ D VW+ +PS + +
Sbjct: 33 DLPEGCVAHILSYICTPEDIVRLSLVSKAFYSAADYDTVWDRFIPSDFSSTISPL----- 197
Query: 89 LSTMSKKDVYTKLC-QPNRFDGGTKEVLLDKYSGQVCLFMSSKSLKITGI----DDRRYW 143
S+ SKKD+Y L +P D G K L+K + + C +S++ + IT + +YW
Sbjct: 198 SSSNSKKDLYFTLSDRPTIIDQGRKSFQLEKRTAKKCYMLSARDISITWAPTQGEASQYW 377
Query: 144 IYIPTEESR 152
+ ESR
Sbjct: 378 EWKSLPESR 404
>BE022522
Length = 414
Score = 58.9 bits (141), Expect = 3e-09
Identities = 25/41 (60%), Positives = 30/41 (72%)
Frame = +2
Query: 239 YYHVGDFMVTKPNKPIKIKFSLAQIDCTHTKGGLCIDSAII 279
+ H G+F+ PN +KIK SL QIDCTHTKGGLC+DS I
Sbjct: 278 HQHAGNFVSKNPNDLMKIKISLTQIDCTHTKGGLCVDSVFI 400
>TC235055
Length = 599
Score = 57.4 bits (137), Expect = 8e-09
Identities = 23/48 (47%), Positives = 34/48 (69%)
Frame = +2
Query: 243 GDFMVTKPNKPIKIKFSLAQIDCTHTKGGLCIDSAIIRPTEFRERLKK 290
G+F V+ +++FS+ QIDCTH+KGGLC+DS I P + RER ++
Sbjct: 107 GEFXVSGSEPTTQVRFSMKQIDCTHSKGGLCVDSVFIVPNDLRERKRR 250
>BQ610780
Length = 447
Score = 52.0 bits (123), Expect = 3e-07
Identities = 36/96 (37%), Positives = 49/96 (50%), Gaps = 1/96 (1%)
Frame = +3
Query: 28 LGDIPESCISSILMHLDPHDICKLARVNRAFHRASSADFVWESKLPSCYKFLANKFLGEE 87
L D+PE CI+ IL + P D C+L+ V+ AF A+ +D VW+ FL + F
Sbjct: 6 LQDLPEGCIAKILSYTTPVDACRLS-VSIAFRSAAESDTVWDC-------FLLSDFTSFI 161
Query: 88 NLSTMSKKDVYTKLCQ-PNRFDGGTKEVLLDKYSGQ 122
S+ SK D+Y L P D G K V L K G+
Sbjct: 162 PPSSTSKNDLYFTLSDLPTIMDQGRKSVQLAKGPGR 269
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.320 0.135 0.414
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 14,230,745
Number of Sequences: 63676
Number of extensions: 217897
Number of successful extensions: 978
Number of sequences better than 10.0: 55
Number of HSP's better than 10.0 without gapping: 954
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 961
length of query: 291
length of database: 12,639,632
effective HSP length: 96
effective length of query: 195
effective length of database: 6,526,736
effective search space: 1272713520
effective search space used: 1272713520
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 58 (26.9 bits)
Medicago: description of AC149494.4


