

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC148819.7 - phase: 0
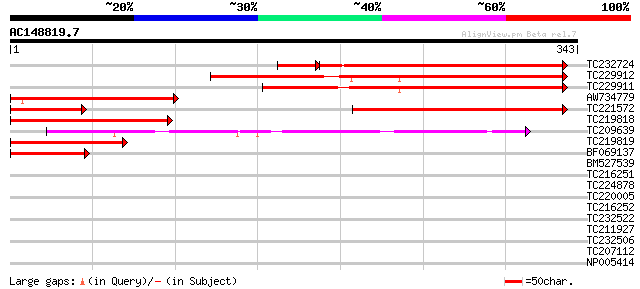
(343 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC232724 277 3e-82
TC229912 similar to UP|Q9FMK9 (Q9FMK9) Emb|CAB76911.1 (At5g63140... 211 5e-55
TC229911 similar to UP|Q9FMK9 (Q9FMK9) Emb|CAB76911.1 (At5g63140... 186 2e-47
AW734779 174 7e-44
TC221572 similar to UP|Q9FMK9 (Q9FMK9) Emb|CAB76911.1 (At5g63140... 162 2e-40
TC219818 similar to UP|Q9FMK9 (Q9FMK9) Emb|CAB76911.1 (At5g63140... 115 4e-26
TC209639 104 5e-23
TC219819 similar to UP|Q9FMK9 (Q9FMK9) Emb|CAB76911.1 (At5g63140... 82 5e-16
BF069137 57 9e-09
BM527539 35 0.049
TC216251 homologue to UP|Q9SMJ5 (Q9SMJ5) DTDP-glucose 4-6-dehydr... 33 0.19
TC224878 32 0.55
TC220005 30 1.2
TC216252 homologue to UP|Q9AV98 (Q9AV98) UDP-D-glucuronate carbo... 30 1.6
TC232522 similar to UP|Q05680 (Q05680) Auxin-responsive GH3 prod... 29 2.7
TC211927 similar to UP|O53882 (O53882) POSSIBLE CONSERVED INTEGR... 29 2.7
TC232506 29 3.5
TC207112 similar to UP|Q6J5M7 (Q6J5M7) Purple acid phosphatase 1... 29 3.5
NP005414 auxin-responsive GH3 product 28 6.0
>TC232724
Length = 786
Score = 277 bits (709), Expect(2) = 3e-82
Identities = 128/150 (85%), Positives = 137/150 (91%)
Frame = +2
Query: 188 QAQEQDPLHSTDHVTSPITPPALAFFHIPIPEVRQLFYKQIVGQFQEGVACSRVNSAVLQ 247
Q Q++DPLH TD + S + PPALAFFHIPIPE+ LFYK+I+GQFQE VACSRVNS VLQ
Sbjct: 155 QGQKRDPLHPTDAI-STMKPPALAFFHIPIPEIPHLFYKEIIGQFQEAVACSRVNSGVLQ 331
Query: 248 TFVSMGDVKAVFIGHDHTNDFCGNLDGIWFCYGGGFGYHGYGKAGWPRRARIILAELQKG 307
FVSMGDVKAVFIGHDHTNDFCGNLDGIWFCYGGGFGYHGYGKAGWPRRARIILAELQKG
Sbjct: 332 AFVSMGDVKAVFIGHDHTNDFCGNLDGIWFCYGGGFGYHGYGKAGWPRRARIILAELQKG 511
Query: 308 KESWTSVQKIMTWKRLDDEKMSKIDEQILW 337
K+SW VQ+IMTWKRLDDEKMSKIDEQILW
Sbjct: 512 KKSWMDVQRIMTWKRLDDEKMSKIDEQILW 601
Score = 46.2 bits (108), Expect(2) = 3e-82
Identities = 20/26 (76%), Positives = 22/26 (83%)
Frame = +1
Query: 163 YQGIRTYDWIKDSQLHWLRHVSQEPQ 188
YQGIRTY WIK+SQL+WLR VS E Q
Sbjct: 1 YQGIRTYGWIKESQLNWLRRVSHEFQ 78
>TC229912 similar to UP|Q9FMK9 (Q9FMK9) Emb|CAB76911.1 (At5g63140), partial
(50%)
Length = 900
Score = 211 bits (536), Expect = 5e-55
Identities = 109/225 (48%), Positives = 136/225 (60%), Gaps = 9/225 (4%)
Frame = +3
Query: 122 KMSKIDGFGNYNLRVYGAPGSMMANSSVLNLFFLDSGDRVVYQGIRTYDWIKDSQLHWLR 181
++ IDGFGNYNL V G G+ N SVLNL+FLDSGD I YDWIK SQ W +
Sbjct: 36 QLHSIDGFGNYNLEVGGVEGTDFENKSVLNLYFLDSGDYSQVSTILGYDWIKPSQQLWFQ 215
Query: 182 HVSQEPQAQEQDPLHSTDHVTSPI----TPPALAFFHIPIPEVRQLFYKQIVGQFQE--- 234
S E + +++ P+ P LA+FHIP+PE L + G E
Sbjct: 216 RTSAELRKA---------YISKPVPQKHAAPGLAYFHIPLPEYASLDSSNMTGVKLEPAG 368
Query: 235 -GVACSRVNSAVLQTFVSMGDVKAVFIGHDHTNDFCGNLDGIWFCYGGGFGYHGYGKAGW 293
G++ VNS T ++ GDVKAVF GHDH NDFCGNL I CY GGFGYH YGKAGW
Sbjct: 369 NGISSPSVNSGFFTTLLAAGDVKAVFTGHDHLNDFCGNLMNIQLCYAGGFGYHAYGKAGW 548
Query: 294 PRRARIILAELQK-GKESWTSVQKIMTWKRLDDEKMSKIDEQILW 337
RRAR+++A L+K K SW V+ I TWKRLDD+ ++ ID ++LW
Sbjct: 549 SRRARVVVASLEKTEKGSWGDVKSIKTWKRLDDQHLTGIDGEVLW 683
>TC229911 similar to UP|Q9FMK9 (Q9FMK9) Emb|CAB76911.1 (At5g63140), partial
(45%)
Length = 778
Score = 186 bits (471), Expect = 2e-47
Identities = 96/190 (50%), Positives = 116/190 (60%), Gaps = 6/190 (3%)
Frame = +1
Query: 154 FLDSGDRVVYQGIRTYDWIKDSQLHWLRHVSQE-PQAQEQDPLHSTDHVTSPITPPALAF 212
FLDSGD I YDWIK SQ W + S E +A P+ D P LA+
Sbjct: 1 FLDSGDYSQVSTILGYDWIKPSQQLWFQRTSAELRKAYISKPVPQKD------AAPGLAY 162
Query: 213 FHIPIPEVRQLFYKQIVGQFQE----GVACSRVNSAVLQTFVSMGDVKAVFIGHDHTNDF 268
FHIP+PE + G QE G++ VNS T ++ GDVKAVF GHDH NDF
Sbjct: 163 FHIPLPEYASFDSSNMTGVKQEPDGNGISSPSVNSGFFTTLLAAGDVKAVFTGHDHINDF 342
Query: 269 CGNLDGIWFCYGGGFGYHGYGKAGWPRRARIILAELQK-GKESWTSVQKIMTWKRLDDEK 327
CGNL I CYGGGFGYH YGKAGWPRRAR+++A L+K GK SW V+ I TWKRLDD+
Sbjct: 343 CGNLMNIQLCYGGGFGYHAYGKAGWPRRARVVVASLEKTGKGSWGDVKSIKTWKRLDDQH 522
Query: 328 MSKIDEQILW 337
++ ID ++LW
Sbjct: 523 LTGIDGEVLW 552
>AW734779
Length = 450
Score = 174 bits (440), Expect = 7e-44
Identities = 83/104 (79%), Positives = 93/104 (88%), Gaps = 2/104 (1%)
Frame = +1
Query: 1 MHFGNG--ITKCRDVLASEFEFCSDLNTTLFLKRVIQDETPDFIAFTGDNIFGPSSHDAA 58
MH+G G +T+C DVLASEFEFCSDLNTT FLKR+I E PDF+AFTGDNIFG SS DAA
Sbjct: 139 MHYGTGTSVTRCXDVLASEFEFCSDLNTTRFLKRIILAENPDFLAFTGDNIFGSSSPDAA 318
Query: 59 ESMFKAFGPAMESGLPWAAILGNHDQESTLNREELMSLISLMDY 102
ES+F+AFGP MESGLPWAA+LGNHDQEST++REELMSLISLMDY
Sbjct: 319 ESLFRAFGPVMESGLPWAAVLGNHDQESTMDREELMSLISLMDY 450
>TC221572 similar to UP|Q9FMK9 (Q9FMK9) Emb|CAB76911.1 (At5g63140), partial
(50%)
Length = 854
Score = 162 bits (410), Expect = 2e-40
Identities = 73/131 (55%), Positives = 86/131 (64%), Gaps = 1/131 (0%)
Frame = +1
Query: 208 PALAFFHIPIPEVRQLFYKQIVGQFQEGVACSRVNSAVLQTFVSMGDVKAVFIGHDHTND 267
P LA+FHIP+PE G QEG++ + VNS T V GDVKAVF GHDH ND
Sbjct: 316 PGLAYFHIPLPEYASFDSSNFTGVKQEGISSASVNSGFFSTLVEAGDVKAVFTGHDHVND 495
Query: 268 FCGNLDGIWFCYGGGFGYHGYGKAGWPRRARIILAELQKGKES-WTSVQKIMTWKRLDDE 326
FCG L GI CY GGFGYH YGKAGW RRAR++L L+K W V+ I TWKRLDD+
Sbjct: 496 FCGELSGIHLCYAGGFGYHAYGKAGWSRRARVVLVSLEKTDNGRWEDVKSIRTWKRLDDQ 675
Query: 327 KMSKIDEQILW 337
++ ID Q+LW
Sbjct: 676 NLTGIDGQVLW 708
Score = 56.6 bits (135), Expect = 2e-08
Identities = 26/47 (55%), Positives = 33/47 (69%), Gaps = 1/47 (2%)
Frame = +3
Query: 1 MHFGNG-ITKCRDVLASEFEFCSDLNTTLFLKRVIQDETPDFIAFTG 46
+HF NG T C DVL S++ CSDLNTT F++R+I E P+ I FTG
Sbjct: 135 LHFANGKTTHCLDVLPSQYASCSDLNTTAFIQRIILSEKPNLIVFTG 275
>TC219818 similar to UP|Q9FMK9 (Q9FMK9) Emb|CAB76911.1 (At5g63140), partial
(26%)
Length = 449
Score = 115 bits (287), Expect = 4e-26
Identities = 54/99 (54%), Positives = 71/99 (71%), Gaps = 1/99 (1%)
Frame = +1
Query: 1 MHFGNG-ITKCRDVLASEFEFCSDLNTTLFLKRVIQDETPDFIAFTGDNIFGPSSHDAAE 59
+HF NG T C DVL S++ CSDLNTT F++R+I E P+ I FTGDNIFG + D A+
Sbjct: 151 LHFANGKTTHCLDVLPSQYASCSDLNTTAFIQRIILSEKPNLIVFTGDNIFGYDASDPAK 330
Query: 60 SMFKAFGPAMESGLPWAAILGNHDQESTLNREELMSLIS 98
SM AF PA+ S +PW A+LGNHDQE +L+RE ++ I+
Sbjct: 331 SMDAAFAPAIASNIPWVAVLGNHDQEGSLSREGVIKYIA 447
>TC209639
Length = 1451
Score = 104 bits (260), Expect = 5e-23
Identities = 90/323 (27%), Positives = 134/323 (40%), Gaps = 30/323 (9%)
Frame = +1
Query: 23 DLNTTLFLKRVIQDETPDFIAFTGDNIFGPSSHDAAESMF--KAFGPAMESGLPWAAILG 80
DLN+ + V+ +E PDF+ + GD I + A S++ +A PA G+PWA++ G
Sbjct: 283 DLNSIRVMSTVLHNENPDFVIYLGDVITANNIMIANASLYWDQATAPARNRGIPWASVFG 462
Query: 81 NHDQESTLNREELMSLISLMDYSVSQINP-SADSLTNSAKGHKMSKIDGFGNYNLRV--- 136
NHD + L +S I P T S G + G G NL
Sbjct: 463 NHDDAAFE--------WPLKWFSAPGIPPIHCPQNTTSYSGEEECSFKGTGRLNLMTNEI 618
Query: 137 --------YGAPGSMMANSS---------------VLNLFFLDSGDRVVYQGIRTYDWIK 173
YG P ++ + S V L+FLDSG G + I
Sbjct: 619 KHNGSFSSYG-PRNLWPSVSNYVLQVSSPNDPQTPVAFLYFLDSG------GGSYPEVIS 777
Query: 174 DSQLHWLRHVSQEPQAQEQDPLHSTDHVTSPITPPALAFFHIPIPEVRQLFYKQIVGQFQ 233
Q+ W R ++E + P H+ S F IP P V + +
Sbjct: 778 SGQVEWFRQKAEEVNPDSRVPEIIFWHIPSTAYKVVAPKFGIPKPCVGSIN--------K 933
Query: 234 EGVACSRVNSAVLQTFVSMGDVKAVFIGHDHTNDFCGNLDGIWFCYGGGFGYHGYGKAGW 293
E VA V + ++ V+ VKA+F+GH+H D+C + +W CY GY GYG W
Sbjct: 934 ETVAAQEVETGMMDLLVNRTSVKAIFVGHNHGLDWCCPYEKLWLCYARHTGYGGYG--DW 1107
Query: 294 PRRARII-LAELQKGKESWTSVQ 315
PR ARI+ + + +SW ++
Sbjct: 1108PRGARILEITQTPFSLQSWIRME 1176
>TC219819 similar to UP|Q9FMK9 (Q9FMK9) Emb|CAB76911.1 (At5g63140), partial
(23%)
Length = 452
Score = 81.6 bits (200), Expect = 5e-16
Identities = 40/72 (55%), Positives = 50/72 (68%), Gaps = 1/72 (1%)
Frame = +2
Query: 1 MHFGNG-ITKCRDVLASEFEFCSDLNTTLFLKRVIQDETPDFIAFTGDNIFGPSSHDAAE 59
+HF NG T C DVL S++ CSDLNTT F++R+I E P+ I FTGDNIFG + D A+
Sbjct: 233 LHFANGKTTHCLDVLPSQYASCSDLNTTAFIQRIILSEKPNLIVFTGDNIFGYDASDPAK 412
Query: 60 SMFKAFGPAMES 71
SM AF PA+ S
Sbjct: 413 SMDAAFAPAIAS 448
>BF069137
Length = 397
Score = 57.4 bits (137), Expect = 9e-09
Identities = 28/49 (57%), Positives = 35/49 (71%), Gaps = 1/49 (2%)
Frame = +3
Query: 1 MHFGNG-ITKCRDVLASEFEFCSDLNTTLFLKRVIQDETPDFIAFTGDN 48
MH+ NG T C +VL S+ CSDLNTT+FL R+I+ E P+ I FTGDN
Sbjct: 249 MHYANGKTTHCLNVLPSQNFSCSDLNTTIFLNRMIKAEKPNLIVFTGDN 395
>BM527539
Length = 421
Score = 35.0 bits (79), Expect = 0.049
Identities = 17/46 (36%), Positives = 26/46 (55%), Gaps = 2/46 (4%)
Frame = +1
Query: 40 DFIAFTGDNIFGPSSHDAAESMF--KAFGPAMESGLPWAAILGNHD 83
DF+ + GD I + A S++ +A PA G+PWA++ GN D
Sbjct: 259 DFVIYLGDVITANNIMIANASLYWDQATAPARNRGIPWASVFGNDD 396
>TC216251 homologue to UP|Q9SMJ5 (Q9SMJ5) DTDP-glucose 4-6-dehydratase ,
partial (98%)
Length = 1378
Score = 33.1 bits (74), Expect = 0.19
Identities = 43/213 (20%), Positives = 82/213 (38%), Gaps = 17/213 (7%)
Frame = +1
Query: 91 EELMSLISLMDYSVSQINPSADSLTNSAKGHKMSKIDGFGNYNLRVYGAPGSMMANSSVL 150
+EL+S++ M S N + T + F N+R+ G+ S ++
Sbjct: 40 QELISVVQRMATDSSNGNGHHQTTTKQPPLPSPLRFSKFFQSNMRILVTGGAGFIGSHLV 219
Query: 151 NLFFLDSGDRVV-----YQGIRTY--DWIKDSQLHWLRHVSQEPQAQEQDPLHSTDHVTS 203
+ + + V+ + G + WI + +RH EP E D ++ H+
Sbjct: 220 DRLMENEKNEVIVADNYFTGSKDNLKKWIGHPRFELIRHDVTEPLLIEVDQIY---HLAC 390
Query: 204 PITPPALAFFHIPIPEVRQLFYKQIVGQFQEGVACSRVNSAVLQTFVSMGDVKAVFIGHD 263
P +P + + + P+ ++ ++G RV + +L T S +V + H
Sbjct: 391 PASP--IFYKYNPVKTIK----TNVIGTLNMLGLAKRVGARILLT--STSEVYGDPLVHP 546
Query: 264 HTNDFCGNLD--GIWFCYGGG--------FGYH 286
+ GN++ G+ CY G F YH
Sbjct: 547 QPESYWGNVNPIGVRSCYDEGKRVAETLMFDYH 645
>TC224878
Length = 1228
Score = 31.6 bits (70), Expect = 0.55
Identities = 18/48 (37%), Positives = 23/48 (47%), Gaps = 4/48 (8%)
Frame = +3
Query: 40 DFIAFTGDNIFG---PSSHDAA-ESMFKAFGPAMESGLPWAAILGNHD 83
DF+ TGDN + S HD A + F A W ++LGNHD
Sbjct: 288 DFVVSTGDNFYDNGLTSDHDNAFQESFTQIYTAKSLQNQWYSVLGNHD 431
>TC220005
Length = 968
Score = 30.4 bits (67), Expect = 1.2
Identities = 21/58 (36%), Positives = 27/58 (46%), Gaps = 1/58 (1%)
Frame = +3
Query: 210 LAFFHIPIPEVR-QLFYKQIVGQFQEGVACSRVNSAVLQTFVSMGDVKAVFIGHDHTN 266
LAFFH+ P V + Q QFQ V +L T ++G V A GH HT+
Sbjct: 294 LAFFHVSFPNVA*TMVVSQHHRQFQRRVFHITSQLLILATGRTVGKVHAPRTGHIHTD 467
>TC216252 homologue to UP|Q9AV98 (Q9AV98) UDP-D-glucuronate carboxy-lyase ,
partial (51%)
Length = 640
Score = 30.0 bits (66), Expect = 1.6
Identities = 29/126 (23%), Positives = 51/126 (40%), Gaps = 10/126 (7%)
Frame = +2
Query: 171 WIKDSQLHWLRHVSQEPQAQEQDPLHSTDHVTSPITPPALAFFHIPIPEVRQLFYKQIVG 230
WI + +RH EP E D ++ H+ P +P + + + P+ ++ ++G
Sbjct: 296 WIGHPRFELIRHDVTEPLTIEVDQIY---HLACPASP--IFYKYNPVKTIK----TNVIG 448
Query: 231 QFQEGVACSRVNSAVLQTFVSMGDVKAVFIGHDHTNDFCGNLD--GIWFCYGGG------ 282
RV + +L T S +V + H + GN++ G+ CY G
Sbjct: 449 TLNMLGLAKRVGARILLT--STSEVYGDPLVHPQPEGYWGNVNPIGVRSCYDEGKRVAET 622
Query: 283 --FGYH 286
F YH
Sbjct: 623 LMFDYH 640
>TC232522 similar to UP|Q05680 (Q05680) Auxin-responsive GH3 product, partial
(55%)
Length = 1006
Score = 29.3 bits (64), Expect = 2.7
Identities = 20/68 (29%), Positives = 28/68 (40%), Gaps = 3/68 (4%)
Frame = +2
Query: 181 RHVSQEPQAQEQDPLHSTDHVTSPITPPALAFFHIPIPEVRQL---FYKQIVGQFQEGVA 237
RH+ + P+ Q P H T H +P T FH R+L +K + G
Sbjct: 269 RHLHRNPKPQNLRPCHQTTHDPNPHTKSRTR*FHCERMLGRKLGPHNHKNLAKHKVSGRD 448
Query: 238 CSRVNSAV 245
C R + AV
Sbjct: 449 CDRCHGAV 472
>TC211927 similar to UP|O53882 (O53882) POSSIBLE CONSERVED INTEGRAL MEMBRANE
PROTEIN, partial (12%)
Length = 692
Score = 29.3 bits (64), Expect = 2.7
Identities = 24/74 (32%), Positives = 37/74 (49%), Gaps = 2/74 (2%)
Frame = +1
Query: 37 ETPDFIAFTGDNIFG--PSSHDAAESMFKAFGPAMESGLPWAAILGNHDQESTLNREELM 94
ET ++ AFT D I G PSS D MF+ I+ + DQ+ R EL+
Sbjct: 160 ETDNYAAFT-DYIKGLSPSSLDMELRMFQ--------------IIDDDDQQEAEKRPELV 294
Query: 95 SLISLMDYSVSQIN 108
S+ L+DY + +++
Sbjct: 295 SIGWLLDYFIHELS 336
>TC232506
Length = 437
Score = 28.9 bits (63), Expect = 3.5
Identities = 17/57 (29%), Positives = 25/57 (43%), Gaps = 5/57 (8%)
Frame = +3
Query: 40 DFIAFTGDNIF-----GPSSHDAAESMFKAFGPAMESGLPWAAILGNHDQESTLNRE 91
DF+ TGDN + G D +S K + A W ++LGNHD + +
Sbjct: 252 DFVISTGDNFYDSGLTGIDDPDFDDSFTKIY-TASSLQKQWYSVLGNHDYRGNVEAQ 419
>TC207112 similar to UP|Q6J5M7 (Q6J5M7) Purple acid phosphatase 1, partial
(80%)
Length = 1200
Score = 28.9 bits (63), Expect = 3.5
Identities = 16/48 (33%), Positives = 20/48 (41%), Gaps = 4/48 (8%)
Frame = +1
Query: 40 DFIAFTGDNIFGPS----SHDAAESMFKAFGPAMESGLPWAAILGNHD 83
DF+ TGDN + A E F A W ++LGNHD
Sbjct: 214 DFVISTGDNFYDDGLTGIDDPAFEISFSKIYTAKSLQKQWYSVLGNHD 357
>NP005414 auxin-responsive GH3 product
Length = 1782
Score = 28.1 bits (61), Expect = 6.0
Identities = 12/34 (35%), Positives = 16/34 (46%)
Frame = +3
Query: 181 RHVSQEPQAQEQDPLHSTDHVTSPITPPALAFFH 214
RH+ + P+ Q P H T H +P T P H
Sbjct: 726 RHLHRNPKPQNH*PCHQTTHDPNPQTRPRTC*IH 827
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.321 0.137 0.427
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 16,360,971
Number of Sequences: 63676
Number of extensions: 230986
Number of successful extensions: 1200
Number of sequences better than 10.0: 38
Number of HSP's better than 10.0 without gapping: 1175
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1190
length of query: 343
length of database: 12,639,632
effective HSP length: 98
effective length of query: 245
effective length of database: 6,399,384
effective search space: 1567849080
effective search space used: 1567849080
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 59 (27.3 bits)
Medicago: description of AC148819.7


