

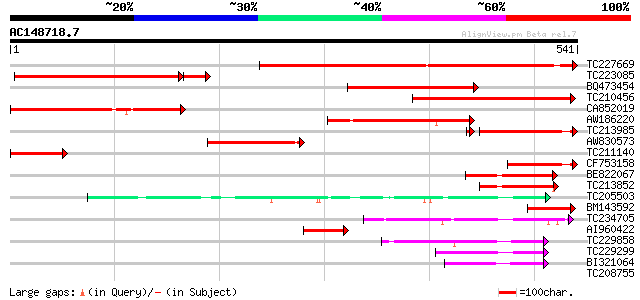
BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC148718.7 + phase: 0
(541 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC227669 homologue to UP|Q9AVQ9 (Q9AVQ9) Phosphate transporter, ... 461 e-130
TC223085 homologue to UP|Q9AVR0 (Q9AVR0) Phosphate transporter, ... 291 2e-85
BQ473454 homologue to GP|24415831|gb phosphate transporter {Medi... 228 5e-60
TC210456 similar to UP|Q9AU01 (Q9AU01) Phosphate transporter 1, ... 221 5e-58
CA852019 homologue to GP|12697486|em phosphate transporter {Sesb... 186 2e-47
AW186220 152 3e-37
TC213985 homologue to UP|Q9AVR0 (Q9AVR0) Phosphate transporter, ... 122 8e-28
AW830573 104 1e-22
TC211140 homologue to UP|Q9AVQ9 (Q9AVQ9) Phosphate transporter, ... 100 3e-21
CF753158 75 1e-13
BE822067 70 2e-12
TC213852 67 2e-11
TC205503 UP|Q7XA50 (Q7XA50) Sorbitol-like transporter, complete 62 6e-10
BM143592 similar to GP|13676622|gb| phosphate transporter 1 {Lup... 60 3e-09
TC234705 weakly similar to GB|CAD70577.1|32698459|MMU549317 solu... 59 4e-09
AI960422 44 2e-04
TC229858 weakly similar to UP|Q8VHD6 (Q8VHD6) Glucose transporte... 44 2e-04
TC229299 similar to UP|GT10_HUMAN (O95528) Solute carrier family... 43 3e-04
BI321064 43 4e-04
TC208755 similar to UP|HXT4_YEAST (P32467) Low-affinity glucose ... 40 0.003
>TC227669 homologue to UP|Q9AVQ9 (Q9AVQ9) Phosphate transporter, partial
(55%)
Length = 1226
Score = 461 bits (1186), Expect = e-130
Identities = 224/303 (73%), Positives = 260/303 (84%)
Frame = +3
Query: 239 TARYTALVAKNAKQAAQDMSSVLQVEIEAEQEKVDKIGVQDKNSFGLFSKEFLRRHGLHL 298
TARYTALVAKNAKQAA DMS VLQVE+EAE++K+ + + +GLFSKEF +RHGLHL
Sbjct: 3 TARYTALVAKNAKQAASDMSKVLQVEVEAEEDKLQHMVESENQKYGLFSKEFAKRHGLHL 182
Query: 299 LGTTSTWFLLDIAYYSSNLFQKDIYSSIGWLPPAQDMNAIHEVFKVARATTLIALCGTVP 358
+GTT TWFLLDIA+YS NLFQKDI+++IGW+PPAQDMNAIHEV+++ARA TLIALC TVP
Sbjct: 183 VGTTVTWFLLDIAFYSQNLFQKDIFTAIGWIPPAQDMNAIHEVYRIARAQTLIALCSTVP 362
Query: 359 GYWFTVAFIDVIGRFAIQLMGFFFMTVFMFALAIPYDHWTKRENRIGFLVMYAMTFFFAN 418
GYWFTVAFID+IGRFAIQLMGFFFMTVFMFALAIPY+HW K N IGF+VMY+ TFFF+N
Sbjct: 363 GYWFTVAFIDIIGRFAIQLMGFFFMTVFMFALAIPYNHW-KNHNNIGFVVMYSFTFFFSN 539
Query: 419 FGPNSTTFVVPAEIFPARLRSTCHGISSAAGKAGAIIGAFGFLYASQSKDPKKRDAGYPA 478
FGPN+TTFVVPAEIFPARLRSTCHGIS+AAGKAGAI+GAFGFLYA+QS +P K D GYP
Sbjct: 540 FGPNATTFVVPAEIFPARLRSTCHGISAAAGKAGAIVGAFGFLYAAQSTNPNKVDHGYPT 719
Query: 479 GIGMKNTLILLAVVNCLGIFFTFLVPEANGKSLEEMSGENEEEEETEAAVVDPQASSNRT 538
GIG+KN+LI+L V+N G+ FT LVPE+ GKSLEE+SGENE+ + A A S RT
Sbjct: 720 GIGVKNSLIVLGVINFFGMVFTLLVPESKGKSLEELSGENED----DGAEAIEMAGSART 887
Query: 539 VPV 541
VPV
Sbjct: 888 VPV 896
>TC223085 homologue to UP|Q9AVR0 (Q9AVR0) Phosphate transporter, partial
(35%)
Length = 634
Score = 291 bits (745), Expect(2) = 2e-85
Identities = 140/162 (86%), Positives = 147/162 (90%)
Frame = +3
Query: 5 QLGVLTALDAAKTQWYHFTAIIIAGMGFFTDAYDLFCIPNVTKLLGRIYYTHPGALKPGT 64
+LGVL ALD AKTQWYHFTAI+IAGMGFFTDAYDLFCI VTKLLGR+YYT PGA KPGT
Sbjct: 66 ELGVLNALDVAKTQWYHFTAIVIAGMGFFTDAYDLFCISLVTKLLGRLYYTEPGAPKPGT 245
Query: 65 LPPNVSAAVNGVALCGTLAGQLFFGWLGDKMGRKKVYGLTLALMVFSSLASGLSFGHSAK 124
LPP V A+ GVALCGTLAGQLFFGWLGDKMGRKKVYGLTL LMV SSLASGLSFG +A+
Sbjct: 246 LPPRVQASATGVALCGTLAGQLFFGWLGDKMGRKKVYGLTLILMVVSSLASGLSFGSTAE 425
Query: 125 GTIGTLCFFRFWLGFGIGGDYPLSATIMSEYANKKTRGAFIA 166
G + TLCFFRFWLGFGIGGDYPLSATIMSEYANKKTRGAFIA
Sbjct: 426 GVMATLCFFRFWLGFGIGGDYPLSATIMSEYANKKTRGAFIA 551
Score = 43.5 bits (101), Expect(2) = 2e-85
Identities = 19/25 (76%), Positives = 24/25 (96%)
Frame = +1
Query: 167 SVFAMQGFGILAGGIVSLVVSTAFD 191
+VFAMQGFGILAGG+V+L+VS A+D
Sbjct: 553 AVFAMQGFGILAGGVVALIVSYAYD 627
>BQ473454 homologue to GP|24415831|gb phosphate transporter {Medicago
sativa}, partial (55%)
Length = 426
Score = 228 bits (581), Expect = 5e-60
Identities = 103/125 (82%), Positives = 119/125 (94%)
Frame = +1
Query: 323 YSSIGWLPPAQDMNAIHEVFKVARATTLIALCGTVPGYWFTVAFIDVIGRFAIQLMGFFF 382
+S+IGW+PPA++MNAIHEV+K+ARA TLIALC TVPGYWFTVA ID +GRFAIQLMGFFF
Sbjct: 52 WSAIGWIPPAKEMNAIHEVYKIARAQTLIALCSTVPGYWFTVALIDYMGRFAIQLMGFFF 231
Query: 383 MTVFMFALAIPYDHWTKRENRIGFLVMYAMTFFFANFGPNSTTFVVPAEIFPARLRSTCH 442
MTVFMFALAIPY HW++++NRIGF+VMY+ TFFFANFGPN+TTFVVPAEIFPARLRSTCH
Sbjct: 232 MTVFMFALAIPYHHWSEKDNRIGFVVMYSFTFFFANFGPNATTFVVPAEIFPARLRSTCH 411
Query: 443 GISSA 447
GIS+A
Sbjct: 412 GISAA 426
>TC210456 similar to UP|Q9AU01 (Q9AU01) Phosphate transporter 1, partial
(29%)
Length = 617
Score = 221 bits (564), Expect = 5e-58
Identities = 109/157 (69%), Positives = 128/157 (81%), Gaps = 1/157 (0%)
Frame = +2
Query: 385 VFMFALAIPYDHWTKRENRIGFLVMYAMTFFFANFGPNSTTFVVPAEIFPARLRSTCHGI 444
V+M ALA+PYDHWT ++NRIGF V+ AMTFFFANFGPN+TTF+VPAEIFPAR RSTCHGI
Sbjct: 2 VYMCALAVPYDHWTHKDNRIGFRVIDAMTFFFANFGPNATTFIVPAEIFPARFRSTCHGI 181
Query: 445 SSAAGKAGAIIGAFGFLYASQSKDPKKRDAGYPAGIGMKNTLILLAVVNCLGIFFTFLVP 504
SS++GK GAI+GAFGFLY +Q+KD K DA YPAGIG+KN LI VVN L +FTFLVP
Sbjct: 182 SSSSGKLGAIVGAFGFLYLAQNKDKSKADARYPAGIGVKNALIA*GVVNILRFYFTFLVP 361
Query: 505 EANGKSLEEMSGENEEEEET-EAAVVDPQASSNRTVP 540
EANGKSLEEMS EN+E+ T E + ++NRTVP
Sbjct: 362 EANGKSLEEMSSENDEDVGTQEESEQSHSHNNNRTVP 472
>CA852019 homologue to GP|12697486|em phosphate transporter {Sesbania
rostrata}, partial (21%)
Length = 793
Score = 186 bits (472), Expect = 2e-47
Identities = 107/170 (62%), Positives = 115/170 (66%), Gaps = 3/170 (1%)
Frame = +2
Query: 1 MAREQLGVLTALDAAKTQWYHFTAIIIAGMGFFTDAYDLFCIPNVTKLLGRIYYTHPGAL 60
MA QLGVL ALD AKTQWYHFTAI+IAGMGFFTDAYDLFCI VTKLLGR+YYT
Sbjct: 296 MAGGQLGVLNALDVAKTQWYHFTAIVIAGMGFFTDAYDLFCISLVTKLLGRLYYTDITNP 475
Query: 61 KPGTLPPNVSAAVNGVALCGTLAGQLFFGWLGDKMGRKKVYGLTLALMVF--SSLASGLS 118
KPG LPPNV AAV GVALCGTLAGQLFFGWLGDK+GRK GL L + S+ G S
Sbjct: 476 KPGVLPPNVQAAVTGVALCGTLAGQLFFGWLGDKLGRK---GLWLNSYAYGGGSVP*G-S 643
Query: 119 FGHSAKGTIG-TLCFFRFWLGFGIGGDYPLSATIMSEYANKKTRGAFIAS 167
+ G G L F LGFG+ G YP E A KKT+G FI S
Sbjct: 644 LLETPPGCHGHPLVSSGFGLGFGLAGTYPFLVQSWLEIATKKTQGHFIGS 793
>AW186220
Length = 425
Score = 152 bits (385), Expect = 3e-37
Identities = 72/143 (50%), Positives = 99/143 (68%), Gaps = 3/143 (2%)
Frame = +1
Query: 304 TWFLLDIAYYSSNLFQKDIYSSIGWLPPAQDMNAIHEVFKVARATTLIALCGTVPGYWFT 363
TWFL+DI +YS LFQ +IY +L +D++ E F A +IA+C T+PGY+F+
Sbjct: 1 TWFLVDIVFYSQVLFQSEIYKR--YLDKKEDVDVYQETFHAAWIQAVIAVCSTIPGYFFS 174
Query: 364 VAFIDVIGRFAIQLMGFFFMTVFMFALAIPY-DHWTKRENRIG--FLVMYAMTFFFANFG 420
+ FID GR +MGFFFM + F++ IPY +WT +E+ F+V+Y + FFFANFG
Sbjct: 175 MYFIDKWGRVKNPMMGFFFMALAFFSIGIPYYSYWTTKEHHKNKVFMVLYGLAFFFANFG 354
Query: 421 PNSTTFVVPAEIFPARLRSTCHG 443
PN+TTF+VPAE+FPAR RS+CHG
Sbjct: 355 PNTTTFIVPAELFPARFRSSCHG 423
>TC213985 homologue to UP|Q9AVR0 (Q9AVR0) Phosphate transporter, partial
(15%)
Length = 583
Score = 122 bits (305), Expect(2) = 8e-28
Identities = 61/93 (65%), Positives = 71/93 (75%)
Frame = +2
Query: 449 GKAGAIIGAFGFLYASQSKDPKKRDAGYPAGIGMKNTLILLAVVNCLGIFFTFLVPEANG 508
GKAGAI+GAFGFLYA+QSKDP K DAGYP GIG+KN+LI+L V+N +G+ FT LVPEA G
Sbjct: 71 GKAGAIVGAFGFLYAAQSKDPSKTDAGYPTGIGIKNSLIMLGVINFIGMLFTLLVPEAKG 250
Query: 509 KSLEEMSGENEEEEETEAAVVDPQASSNRTVPV 541
KSLEE+SGEN E + A S RTVPV
Sbjct: 251 KSLEELSGENNENDAEHAV-------SARTVPV 328
Score = 20.4 bits (41), Expect(2) = 8e-28
Identities = 7/7 (100%), Positives = 7/7 (100%)
Frame = +3
Query: 437 LRSTCHG 443
LRSTCHG
Sbjct: 33 LRSTCHG 53
>AW830573
Length = 304
Score = 104 bits (259), Expect = 1e-22
Identities = 54/93 (58%), Positives = 64/93 (68%)
Frame = +2
Query: 189 AFDHAFKAPPYKVDAAASLVPEADYVWRIILMFGAVPAGLTYYWRMKMPETARYTALVAK 248
++DH FK P Y+ ASL P Y WRI+ GA PA LTYYWRM MPET RYT LVAK
Sbjct: 17 SYDHKFKLPSYEQHPYASLDPAFVYDWRILTKAGA*PAALTYYWRMNMPETPRYTLLVAK 196
Query: 249 NAKQAAQDMSSVLQVEIEAEQEKVDKIGVQDKN 281
AKQAA DMS VL +E+EAE E V KI ++++
Sbjct: 197 TAKQAAADMSGVLHLELEAEDENV-KISTENES 292
>TC211140 homologue to UP|Q9AVQ9 (Q9AVQ9) Phosphate transporter, partial
(10%)
Length = 466
Score = 99.8 bits (247), Expect = 3e-21
Identities = 47/55 (85%), Positives = 49/55 (88%)
Frame = +1
Query: 1 MAREQLGVLTALDAAKTQWYHFTAIIIAGMGFFTDAYDLFCIPNVTKLLGRIYYT 55
MA QLGVL ALD AKTQWYHFTAI+IAGMGFFTDAYDLFCI VTKLLGR+YYT
Sbjct: 292 MAGGQLGVLNALDVAKTQWYHFTAIVIAGMGFFTDAYDLFCISLVTKLLGRLYYT 456
>CF753158
Length = 473
Score = 74.7 bits (182), Expect = 1e-13
Identities = 38/66 (57%), Positives = 46/66 (69%)
Frame = -3
Query: 476 YPAGIGMKNTLILLAVVNCLGIFFTFLVPEANGKSLEEMSGENEEEEETEAAVVDPQASS 535
YP GIG+KN+LI+L V+N +G+ FT LVPEA GKSLEE+SGEN E + A S
Sbjct: 387 YPTGIGIKNSLIMLGVINFIGMLFTLLVPEAKGKSLEELSGENNENDAEHAV-------S 229
Query: 536 NRTVPV 541
RTVPV
Sbjct: 228 ARTVPV 211
>BE822067
Length = 610
Score = 70.1 bits (170), Expect = 2e-12
Identities = 41/88 (46%), Positives = 54/88 (60%), Gaps = 1/88 (1%)
Frame = -1
Query: 436 RLRSTCHGISSAAGKAGAIIGAFGFLYASQSKDPKKRDAGYPAGIGMKNTLILLAVVNCL 495
R RS+CHGIS GAIIG+ GFL+AS +K++ GY IGMK +LI+L V L
Sbjct: 610 RFRSSCHGISGXVXXVGAIIGSVGFLWASH----RKKEDGYXKXIGMKVSLIILGGVCLL 443
Query: 496 GIFFT-FLVPEANGKSLEEMSGENEEEE 522
G+ T F E G+SL+E E EE++
Sbjct: 442 GMVITYFFTRETMGRSLQENEVEIEEQD 359
>TC213852
Length = 411
Score = 67.0 bits (162), Expect = 2e-11
Identities = 42/78 (53%), Positives = 51/78 (64%), Gaps = 3/78 (3%)
Frame = +3
Query: 449 GKAGAIIGAFGFLYASQSKDPKKRDAGYPAGIGMKNTLILLAVVNCLGIFFTFL-VPEAN 507
GK GAIIG+ GFL+AS KK++ GYP GIGM+ TLI+L VV LG+ T+L E
Sbjct: 36 GKVGAIIGSVGFLWASH----KKKENGYPKGIGMEVTLIILGVVCLLGMLVTYLFTRETM 203
Query: 508 GKSLEEMSGE--NEEEEE 523
G+SLEE E N E EE
Sbjct: 204 GRSLEENEVESVNHEVEE 257
>TC205503 UP|Q7XA50 (Q7XA50) Sorbitol-like transporter, complete
Length = 1928
Score = 62.0 bits (149), Expect = 6e-10
Identities = 109/476 (22%), Positives = 183/476 (37%), Gaps = 34/476 (7%)
Frame = +1
Query: 75 GVALCGTLAGQLFFGWLGDKMGRKKVYGLTLALMVFSSLASGLSFGHSAKGTIGTLCFFR 134
G+ +L G G D +GR+ GL A+ + S G +S L R
Sbjct: 388 GIINLYSLIGSCLAGRTSDWIGRRYTIGLGGAIFLVGSTLMGFYPHYSF------LMCGR 549
Query: 135 FWLGFGIGGDYPLSATIMSEYANKKTRGAFIASVFAMQGFGILAGGIVSLVVSTAFDHAF 194
F G GIG ++ +E + +RG + GIL G I + + F
Sbjct: 550 FVAGIGIGYALMIAPVYTAEVSPASSRGFLTSFPEVFINGGILLGYISN--------YGF 705
Query: 195 KAPPYKVDAAASLVPEADYVWRIILMFGAVPAGLTYYWRMKMPETARYTALVAK------ 248
KV WR++L GA+P+ + + MPE+ R+ + +
Sbjct: 706 SKLTLKVG------------WRMMLGVGAIPSVVLTEGVLAMPESPRWLVMRGRLGEARK 849
Query: 249 ------NAKQAAQ-DMSSVLQVEIEAEQEKVDKIGV-QDKNSFGLFSKEFLR-----RH- 294
++K+ AQ ++ + Q E D + V + N G++ + FL RH
Sbjct: 850 VLNKTSDSKEEAQLRLAEIKQAAGIPESCNDDVVQVNKQSNGEGVWKELFLYPTPAIRHI 1029
Query: 295 -----GLHLLGTTSTWFLLDIAYYSSNLFQKDIYSSIGWLPPAQDMNAIHEVFKVARATT 349
G+H S + + YS +F+K ++ D + + V T
Sbjct: 1030VIAALGIHFFQQASG--VDAVVLYSPRIFEKAGITN--------DTHKLLATVAVGFVKT 1179
Query: 350 LIALCGTVPGYWFTVAFIDVIGRFAIQLMGFFFMTVFMFALAIPY---DHWTKR-----E 401
+ L T FT +D +GR + L M + + LAI DH ++
Sbjct: 1180VFILAAT-----FT---LDRVGRRPLLLSSVGGMVLSLLTLAISLTVIDHSERKLMWAVG 1335
Query: 402 NRIGFLVMYAMTFFFANFGPNSTTFVVPAEIFPARLRSTCHGISSAAGKAGAIIGAFGFL 461
+ I ++ Y TF + G T+V +EIFP RLR+ A + + + + FL
Sbjct: 1336SSIAMVLAYVATF---SIGAGPITWVYSSEIFPLRLRAQGAAAGVAVNRTTSAVVSMTFL 1506
Query: 462 YASQSKDPKKRDAGYPAGIGMKNTLILLAVVNCLG-IFFTFLVPEANGKSLEEMSG 516
+++ I + L + +G IFF ++PE GK+LE+M G
Sbjct: 1507SLTRA-------------ITIGGAFFLYCGIATVGWIFFYTVLPETRGKTLEDMEG 1635
>BM143592 similar to GP|13676622|gb| phosphate transporter 1 {Lupinus albus},
partial (9%)
Length = 378
Score = 59.7 bits (143), Expect = 3e-09
Identities = 31/47 (65%), Positives = 36/47 (75%), Gaps = 1/47 (2%)
Frame = -1
Query: 495 LGIFFTFLVPEANGKSLEEMSGENEEEEET-EAAVVDPQASSNRTVP 540
LG FFTFLVPEANGKSLEEMSGEN+E+ T E + ++NRTVP
Sbjct: 375 LGFFFTFLVPEANGKSLEEMSGENDEDVGTQEESEQSHSQNNNRTVP 235
>TC234705 weakly similar to GB|CAD70577.1|32698459|MMU549317 solute carrier
family 2 (facilitated glucose transporter), member 12
{Mus musculus;} , partial (5%)
Length = 992
Score = 59.3 bits (142), Expect = 4e-09
Identities = 57/210 (27%), Positives = 93/210 (44%), Gaps = 9/210 (4%)
Frame = +1
Query: 338 IHEVFKVARATTLIALCGTVPGYWFTVAFIDVIGRFAIQLMGFFFMTVFMFALAIPYDHW 397
I + K+ AT + + T+ + ID +GR + ++ MTV +F +
Sbjct: 25 IEDNSKLLAATVAVGVAKTI-FILVAIILIDKLGRKPLLMISTIGMTVCLFCMGATLALL 201
Query: 398 TKRENRIGFLVMYA---MTFFFANFGPNSTTFVVPAEIFPARLRSTCHGISSAAGKAGAI 454
K I +++ + FF GP +V+ +EIFP R+R+ + + A + +
Sbjct: 202 GKGSFAIALAILFVCGNVAFFSVGLGP--VCWVLTSEIFPLRVRAQASALGAVANRVCSG 375
Query: 455 IGAFGFLYASQSKDPKKRDAGYPAGIGMKNTLILLAVVNCLGIFFTF-LVPEANGKSLE- 512
+ A FL S++ I + T + A ++ L I F LVPE GKSLE
Sbjct: 376 LVAMSFLSVSEA-------------ISVAGTFFVFAAISALAIAFVVTLVPETKGKSLEQ 516
Query: 513 -EMSGENEEE---EETEAAVVDPQASSNRT 538
EM +NE E +E E V+ Q N+T
Sbjct: 517 IEMMFQNEYEIQGKEMELGDVE-QLVQNKT 603
>AI960422
Length = 130
Score = 43.5 bits (101), Expect = 2e-04
Identities = 19/43 (44%), Positives = 28/43 (64%)
Frame = +2
Query: 281 NSFGLFSKEFLRRHGLHLLGTTSTWFLLDIAYYSSNLFQKDIY 323
+++ L S E+LR+HG L + TWFL+DIA YS L ++Y
Sbjct: 2 HAYPLLSWEYLRKHGPDLFACSFTWFLVDIAVYSQVLVHSELY 130
>TC229858 weakly similar to UP|Q8VHD6 (Q8VHD6) Glucose transporter 10 (Mus
musculus 13 days embryo heart cDNA, RIKEN full-length
enriched library, clone:D330001H08 product:solute
carrier family 2 (facilitated glucose transporter),
member 10, full insert sequence), partial (4%)
Length = 715
Score = 43.5 bits (101), Expect = 2e-04
Identities = 37/164 (22%), Positives = 70/164 (42%), Gaps = 4/164 (2%)
Frame = +3
Query: 355 GTVPGYWFTVAFIDVIGRFAIQLMGFFFMTVFMFALAIPYDHWTKRENRI-GFLVMYAMT 413
GT+ G + ID GR + L + V + LA + + N + G+L + +
Sbjct: 171 GTILGIYL----IDHAGRKKLALSSLGGVIVSLVILAFAFYKQSSTSNELYGWLAVVGLA 338
Query: 414 FFFANFGPNS--TTFVVPAEIFPARLRSTCHGISSAAGKAGAIIGAFGFLYASQSKDPKK 471
+ F P + + +EI+P R C G+S+ +I + FL ++
Sbjct: 339 LYIGFFSPGMGPVPWTLSSEIYPEEYRGICGGMSATVCWVSNLIVSETFLSIAE------ 500
Query: 472 RDAGYPAGIGMKNTLILLAVVNCLGIFFTFL-VPEANGKSLEEM 514
GIG+ +T +++ V+ + F + VPE G + +E+
Sbjct: 501 -------GIGIGSTFLIIGVIAVVAFVFVLVYVPETKGLTFDEV 611
>TC229299 similar to UP|GT10_HUMAN (O95528) Solute carrier family 2,
facilitated glucose transporter, member 10 (Glucose
transporter type 10), partial (6%)
Length = 1060
Score = 43.1 bits (100), Expect = 3e-04
Identities = 30/109 (27%), Positives = 51/109 (46%), Gaps = 1/109 (0%)
Frame = +2
Query: 407 LVMYAMTFFFANFGPNSTTFVVPAEIFPARLRSTCHGISSAAGKAGAIIGAFGFLYASQS 466
L+ A+ F + G + +VV +EI+P R R C GI+S +I + FL +++
Sbjct: 533 LIGLALYIIFFSPGMGTVPWVVNSEIYPLRYRGVCGGIASTTVWISNLIVSESFLSLTKA 712
Query: 467 KDPKKRDAGYPAGIGMKNTLILLAVVNCLGIFFTFL-VPEANGKSLEEM 514
+G T ++ +V + IFF + VPE G +EE+
Sbjct: 713 -------------LGTAWTFMMFGIVAIVAIFFVIIFVPETKGVPMEEV 820
>BI321064
Length = 284
Score = 42.7 bits (99), Expect = 4e-04
Identities = 31/100 (31%), Positives = 46/100 (46%), Gaps = 1/100 (1%)
Frame = +3
Query: 416 FANFGPNSTTFVVPAEIFPARLRSTCHGISSAAGKAGAIIGAFGFLYASQSKDPKKRDAG 475
F + G ++ +VV +EI+P R R C GI+S +I + FL +
Sbjct: 9 FFSPGMDTVPWVVNSEIYPLRYRGVCGGIASTTCWVSNLIVSQSFLTLT----------- 155
Query: 476 YPAGIGMKNTLILLAVVNCLGIFFTFL-VPEANGKSLEEM 514
IG T +L V +GIFF + VPE G +EE+
Sbjct: 156 --VAIGTAWTFMLFGFVALIGIFFVLIFVPETKGVPVEEV 269
>TC208755 similar to UP|HXT4_YEAST (P32467) Low-affinity glucose transporter
HXT4 (Low-affinity glucose transporter LGT1), partial
(3%)
Length = 701
Score = 39.7 bits (91), Expect = 0.003
Identities = 26/113 (23%), Positives = 48/113 (42%), Gaps = 3/113 (2%)
Frame = +3
Query: 405 GFLVMYAMTFFFANFGPNS--TTFVVPAEIFPARLRSTCHGISSAAGKAGAIIGAFGFLY 462
G+L + + + A F P + V +E++P R C G+S+ +I FL
Sbjct: 21 GWLAILGLALYIAFFSPGMGPVPWTVNSEVYPEEYRGICGGMSATVNWVSNLIVVQSFLS 200
Query: 463 ASQSKDPKKRDAGYPAGIGMKNTLILLAVVNCLGIFFTFL-VPEANGKSLEEM 514
+ A +G T +++A++ L F + VPE G + +E+
Sbjct: 201 VA-------------AAVGTGPTFLIIAIIAVLAFMFVVVYVPETKGLTFDEV 320
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.325 0.139 0.424
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 22,873,742
Number of Sequences: 63676
Number of extensions: 301117
Number of successful extensions: 2059
Number of sequences better than 10.0: 73
Number of HSP's better than 10.0 without gapping: 2029
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 2045
length of query: 541
length of database: 12,639,632
effective HSP length: 102
effective length of query: 439
effective length of database: 6,144,680
effective search space: 2697514520
effective search space used: 2697514520
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 15 ( 7.0 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.6 bits)
S2: 61 (28.1 bits)
Medicago: description of AC148718.7


