

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC148651.2 - phase: 0
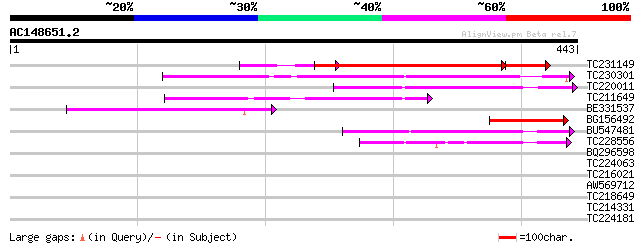
(443 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC231149 similar to GB|BAB10122.1|10176892|AB011475 Na+-dependen... 278 8e-86
TC230301 143 2e-34
TC220011 homologue to GB|AAP78931.1|32306495|BT009663 At4g00370 ... 143 2e-34
TC211649 similar to GB|AAP78931.1|32306495|BT009663 At4g00370 {A... 140 1e-33
BE331537 106 2e-23
BG156492 similar to GP|10176892|dbj Na+-dependent inorganic phos... 100 1e-21
BU547481 89 4e-18
TC228556 weakly similar to UP|Q7PWJ2 (Q7PWJ2) ENSANGP00000021389... 72 5e-13
BQ296598 41 0.001
TC224063 similar to UP|Q948P0 (Q948P0) Aspartic proteinase 2, pa... 34 0.12
TC216021 homologue to UP|Q948P0 (Q948P0) Aspartic proteinase 2, ... 30 2.8
AW569712 29 4.9
TC218649 similar to UP|Q39416 (Q39416) Integral membrane protein... 28 6.3
TC214331 similar to UP|Q7NLY3 (Q7NLY3) Glr0986 protein, partial ... 28 8.3
TC224181 28 8.3
>TC231149 similar to GB|BAB10122.1|10176892|AB011475 Na+-dependent inorganic
phosphate cotransporter-like protein {Arabidopsis
thaliana;} , partial (42%)
Length = 707
Score = 278 bits (712), Expect(2) = 8e-86
Identities = 134/150 (89%), Positives = 145/150 (96%)
Frame = +3
Query: 239 SLPVWAIVVNNFTFHYALYVLMNWLPTYFELGLKLSLHEMGSSKMLPYLNMFVFSNIGGV 298
S PVWAIVVNNFTFHYALYVLMNWLPTYFELGL+LSL +MGSSKM+PYLNMF+FSNIGGV
Sbjct: 147 SFPVWAIVVNNFTFHYALYVLMNWLPTYFELGLQLSLQDMGSSKMMPYLNMFLFSNIGGV 326
Query: 299 VADYLITRRVMSVTRTRKFLNTIGFLVASFALIVIPSFRTSGGAVFCSSVALGFLALGRA 358
VADYLITRR++SVT+TRKFLNT+GFLVAS AL++IPSFRTSGGAVFCSSVALGFLALGRA
Sbjct: 327 VADYLITRRILSVTKTRKFLNTVGFLVASLALVIIPSFRTSGGAVFCSSVALGFLALGRA 506
Query: 359 GFAVNHMDVAPRYAGIVMGVSNTAGTLAGI 388
GFA NHMD+APRYAGIVMGVSNTAGTL GI
Sbjct: 507 GFAGNHMDIAPRYAGIVMGVSNTAGTLTGI 596
Score = 57.4 bits (137), Expect(2) = 8e-86
Identities = 27/35 (77%), Positives = 31/35 (88%)
Frame = +2
Query: 388 IVGVDLTGKLLEAAKASDSDLSTPESWRLVFFIPG 422
++GVDLTGKL EAAKA+ SDLS+PESWR VF IPG
Sbjct: 596 LLGVDLTGKLPEAAKAAKSDLSSPESWRAVFSIPG 700
Score = 54.7 bits (130), Expect = 8e-08
Identities = 29/79 (36%), Positives = 42/79 (52%)
Frame = +1
Query: 180 IWFRYSSDPKSSASGAGESLLPVNKKIDRRVSDLKVGVEKNGGEGKKSGVGIPWVKILTS 239
+WF+Y++DPKS+ASG GES+LPVNKKID K IPWVK +
Sbjct: 7 LWFKYATDPKSTASGVGESVLPVNKKIDTH-------------NKKPLSAKIPWVKXFNT 147
Query: 240 LPVWAIVVNNFTFHYALYV 258
+ +++ + LY+
Sbjct: 148 ASLCGRLLSTISPFTMLYM 204
>TC230301
Length = 1398
Score = 143 bits (360), Expect = 2e-34
Identities = 99/329 (30%), Positives = 154/329 (46%), Gaps = 7/329 (2%)
Frame = +1
Query: 120 HTVLAQWVPPHERSRSVSLTTSGMYLGAAFGMLFLPSLVKFKGPQSVFVAESFLGFVWCF 179
H +W P ERSR+V ++ +G LG A G+ P L+ G FV GF+W
Sbjct: 94 HGCQVRWFPQTERSRAVGISMAGFQLGCAIGLTLSPILMSQGGIXGPFVIFGLSGFLWVL 273
Query: 180 IWFRY-SSDPKSSASGAGESLLPVNKKIDRRVSDLKVGVEKNGGEGKKSGVGIPWVKILT 238
+W SS P S + L ++ +RR V K KK V P+ ++L+
Sbjct: 274 VWLSATSSTPDRSPQISKYELEYIS---NRRHKSFSVETAKP----KKVKVIPPFRRLLS 432
Query: 239 SLPVWAIVVNNFTFHYALYVLMNWLPTYFELGLKLSLHEMGSSKMLPYLNMFVFSNIGGV 298
P W++++ N + + +++W+P YF ++ L +P+ M V G+
Sbjct: 433 KRPTWSLIIANSMHSWGFFTVLSWMPIYFSSVYRVDLRHAAWFSAVPWAVMAVTGYFAGL 612
Query: 299 VADYLITRRVMSVTRTRKFLNTIGFLVASFALIVIPSFRTSGGAVFCSSVALGFLALGRA 358
+D +I SVT TRK + +IGF+ LI + + + ++A G + +
Sbjct: 613 WSDMMIQSGT-SVTLTRKIMQSIGFIGPGLCLIGLATAKNPSIGSAGLTLAFGLKSFSHS 789
Query: 359 GFAVNHMDVAPRYAGIVMGVSNTAGTLAGIVGVDLTGKLLEAAKASDSDLSTPESWRLVF 418
GF VN ++AP+Y+G++ G+SNTAGTLA I G G +E LV
Sbjct: 790 GFLVNLQEIAPQYSGVLHGISNTAGTLAAIFGTVGAGLFVE----------------LVG 921
Query: 419 FIPGLLCVFSSLVFL------LFSTGERI 441
PG L + S L FL LF+TGER+
Sbjct: 922 SFPGFLLLTSLLYFLAAIFYCLFATGERV 1008
>TC220011 homologue to GB|AAP78931.1|32306495|BT009663 At4g00370 {Arabidopsis
thaliana;} , partial (33%)
Length = 904
Score = 143 bits (360), Expect = 2e-34
Identities = 75/190 (39%), Positives = 114/190 (59%)
Frame = +1
Query: 254 YALYVLMNWLPTYFELGLKLSLHEMGSSKMLPYLNMFVFSNIGGVVADYLITRRVMSVTR 313
+ ++L+ W+PTY+ LK +L E G +LP+L M +F+NIGG +AD L+++ +S+T
Sbjct: 1 WGTFILLTWMPTYYNQVLKFNLTESGLFCVLPWLTMAIFANIGGWIADTLVSKG-LSITS 177
Query: 314 TRKFLNTIGFLVASFALIVIPSFRTSGGAVFCSSVALGFLALGRAGFAVNHMDVAPRYAG 373
RK + +IGFL +F L + RT AV C + + G A ++G NH D+ PRYAG
Sbjct: 178 VRKIMQSIGFLGPAFFLTQLSHVRTPAMAVLCMACSQGSDAFSQSGLYSNHQDIGPRYAG 357
Query: 374 IVMGVSNTAGTLAGIVGVDLTGKLLEAAKASDSDLSTPESWRLVFFIPGLLCVFSSLVFL 433
+++G+SNTAG LAG+ G TG +L+ SW VF + L + +LV+
Sbjct: 358 VLLGLSNTAGVLAGVFGTAATGYILQRG-----------SWDDVFKVAVALYIIGTLVWN 504
Query: 434 LFSTGERIFD 443
+FSTGE+I D
Sbjct: 505 IFSTGEKILD 534
>TC211649 similar to GB|AAP78931.1|32306495|BT009663 At4g00370 {Arabidopsis
thaliana;} , partial (35%)
Length = 606
Score = 140 bits (352), Expect = 1e-33
Identities = 80/209 (38%), Positives = 119/209 (56%)
Frame = +2
Query: 122 VLAQWVPPHERSRSVSLTTSGMYLGAAFGMLFLPSLVKFKGPQSVFVAESFLGFVWCFIW 181
+L++WVP ERSRS++L SGMYLG+ G+ F P L+ G SVF + LG VWC +W
Sbjct: 2 ILSKWVPVSERSRSLALVYSGMYLGSVTGLAFSPFLIHQFGWPSVFYSFGSLGTVWCSVW 181
Query: 182 FRYSSDPKSSASGAGESLLPVNKKIDRRVSDLKVGVEKNGGEGKKSGVGIPWVKILTSLP 241
+ S+ L P KK+ K V+ IPW IL+ P
Sbjct: 182 LSKAH----SSPLEDPELRPEEKKLITANCSSKEPVKT-----------IPWRLILSKPP 316
Query: 242 VWAIVVNNFTFHYALYVLMNWLPTYFELGLKLSLHEMGSSKMLPYLNMFVFSNIGGVVAD 301
VWA++V++F ++ ++L+ W+PTY+ LK +L E G +LP+ M + +N+GG +AD
Sbjct: 317 VWALIVSHFCHNWGTFILLTWMPTYYNQVLKFNLTESGLFCVLPWFIMAISANVGGWIAD 496
Query: 302 YLITRRVMSVTRTRKFLNTIGFLVASFAL 330
L+++ +SVTR RK + TIGFL +F L
Sbjct: 497 TLVSKG-LSVTRVRKIMQTIGFLGPAFFL 580
>BE331537
Length = 628
Score = 106 bits (264), Expect = 2e-23
Identities = 57/167 (34%), Positives = 93/167 (55%), Gaps = 3/167 (1%)
Frame = +2
Query: 45 TKGTILSTFYYGYACSQVPGGYFAQKIGGRKILLFSFLLWSLTCALLPLDPNKTL-ILVT 103
T G I S+ +GY +Q+ GG +A K+GG+ +L F + WS+ L P+ L L+
Sbjct: 2 TFGLIQSSL*WGYLLTQIIGGIWADKLGGKLVLGFGVVWWSIATGLTPIAAKLGLPCLLI 181
Query: 104 ARLLVGVAQGFIFPSIHTVLAQWVPPHERSRSVSLTTSGMYLGAAFGMLFLPSLVKFKGP 163
R +G+ +G P+++ +L++W+P ERSRS++L S MYLG+ G+ F P L++ G
Sbjct: 182 MRAFMGIGEGVAMPAMNNILSKWIPVSERSRSLALVYSDMYLGSVTGLAFSPILIQKFGW 361
Query: 164 QSVFVAESFLGFVWCFIWF--RYSSDPKSSASGAGESLLPVNKKIDR 208
SVF + LG +W +W YSS + GA E + + + +
Sbjct: 362 PSVFYSYGSLGSIWFVVWLG*AYSSPNQDPDLGA*EEISILRSNVSK 502
>BG156492 similar to GP|10176892|dbj Na+-dependent inorganic phosphate
cotransporter-like protein {Arabidopsis thaliana},
partial (14%)
Length = 184
Score = 100 bits (249), Expect = 1e-21
Identities = 51/61 (83%), Positives = 55/61 (89%)
Frame = +2
Query: 376 MGVSNTAGTLAGIVGVDLTGKLLEAAKASDSDLSTPESWRLVFFIPGLLCVFSSLVFLLF 435
MGVSNTAGTLAGIVGVDLTGKLLEAAKA++SDLSTPESW +V FIPG LCV +S V LLF
Sbjct: 2 MGVSNTAGTLAGIVGVDLTGKLLEAAKAANSDLSTPESWTIVSFIPGFLCVSTSSVSLLF 181
Query: 436 S 436
S
Sbjct: 182 S 184
>BU547481
Length = 598
Score = 89.0 bits (219), Expect = 4e-18
Identities = 53/181 (29%), Positives = 93/181 (51%)
Frame = -3
Query: 261 NWLPTYFELGLKLSLHEMGSSKMLPYLNMFVFSNIGGVVADYLITRRVMSVTRTRKFLNT 320
+W+P YF+ ++L + +P+ M + + GV +D+LI ++ RKF+ T
Sbjct: 596 SWMPVYFKSVYNVNLKQAAWFSAVPWATMAMSGXLAGVASDFLINAGYPTIF-VRKFMQT 420
Query: 321 IGFLVASFALIVIPSFRTSGGAVFCSSVALGFLALGRAGFAVNHMDVAPRYAGIVMGVSN 380
IGF+ + L+ + T A ++AL + +AGF +N D+AP+YAGI+ G+SN
Sbjct: 419 IGFIGPAVTLLCLNYANTPAVAATLMTIALSLSSFSQAGFMLNIQDIAPQYAGILHGISN 240
Query: 381 TAGTLAGIVGVDLTGKLLEAAKASDSDLSTPESWRLVFFIPGLLCVFSSLVFLLFSTGER 440
AGT+A I+ TG ++ S++ I L +++ + LF+T E+
Sbjct: 239 CAGTIAAIISTIGTGYFVQWL----------GSFQAFLTITACLYFVTTIFWNLFATSEQ 90
Query: 441 I 441
I
Sbjct: 89 I 87
>TC228556 weakly similar to UP|Q7PWJ2 (Q7PWJ2) ENSANGP00000021389, partial
(6%)
Length = 897
Score = 72.0 bits (175), Expect = 5e-13
Identities = 53/173 (30%), Positives = 86/173 (49%), Gaps = 7/173 (4%)
Frame = +2
Query: 274 SLHEMGSSKMLPYLNMFVFSNIGGVVADYLITRRVMSVTRTRKFLNTIGFLVASFALIV- 332
+L E +LP L +++ +AD LI+R V T RK +I FL + + +
Sbjct: 2 NLTEAAWVSILPPLASIFVTSLAAQLADNLISRGV-ETTVVRKICQSIAFLSPAICMTLS 178
Query: 333 -----IPSFRTSGGAVFCSSVALGFLALGRAGFAVNHMDVAPRYAGIVMGVSNTAGTLAG 387
+P + G + S +AL AL +G H D++P YA I++G++NT G + G
Sbjct: 179 SLDLGLPPWEIVG--ILTSGLALSSFAL--SGLYCTHQDMSPEYASILLGITNTVGAIPG 346
Query: 388 IVGVDLTGKLLEAAKASDSDLSTPESWRLVFFIPGL-LCVFSSLVFLLFSTGE 439
IVGV LTG LL++ SW + F P + V ++V+L F++ +
Sbjct: 347 IVGVALTGYLLDST----------HSWSISLFAPSIFFYVTGTIVWLAFASSK 475
>BQ296598
Length = 421
Score = 41.2 bits (95), Expect = 0.001
Identities = 14/37 (37%), Positives = 26/37 (69%)
Frame = +2
Query: 231 IPWVKILTSLPVWAIVVNNFTFHYALYVLMNWLPTYF 267
IPW IL+ PV A+++++F + ++L+ W+PTY+
Sbjct: 149 IPWKLILSKAPVLALIISHFCHN*RTFILLTWMPTYY 259
Score = 32.7 bits (73), Expect = 0.34
Identities = 24/74 (32%), Positives = 40/74 (53%), Gaps = 1/74 (1%)
Frame = +1
Query: 267 FELGLKLSLHEMGSSKMLPYL-NMFVFSNIGGVVADYLITRRVMSVTRTRKFLNTIGFLV 325
+ L L L ++ SS + YL +F+NIGG +AD L+ R +S T R+ + +I FL
Sbjct: 193 YNLPLLSQLKDIYSSNLDAYLLQSGIFANIGGWIADTLV-RNGISTTVIRQIMQSIRFLG 369
Query: 326 ASFALIVIPSFRTS 339
+ L + R++
Sbjct: 370 PAIFLSQLSHVRST 411
>TC224063 similar to UP|Q948P0 (Q948P0) Aspartic proteinase 2, partial (25%)
Length = 519
Score = 34.3 bits (77), Expect = 0.12
Identities = 15/39 (38%), Positives = 21/39 (53%)
Frame = +3
Query: 54 YYGYACSQVPGGYFAQKIGGRKILLFSFLLWSLTCALLP 92
YY +C+ + + G K L F LW+LTC+LLP
Sbjct: 84 YYAPSCTSICSSLRFSLMMGHKYLWLVFCLWALTCSLLP 200
>TC216021 homologue to UP|Q948P0 (Q948P0) Aspartic proteinase 2, complete
Length = 1820
Score = 29.6 bits (65), Expect = 2.8
Identities = 12/20 (60%), Positives = 16/20 (80%)
Frame = +1
Query: 73 GRKILLFSFLLWSLTCALLP 92
G+K L+ F LW+LTC+LLP
Sbjct: 106 GQKHLVTVFCLWALTCSLLP 165
>AW569712
Length = 413
Score = 28.9 bits (63), Expect = 4.9
Identities = 21/65 (32%), Positives = 30/65 (45%)
Frame = -2
Query: 86 LTCALLPLDPNKTLILVTARLLVGVAQGFIFPSIHTVLAQWVPPHERSRSVSLTTSGMYL 145
L C PLDPNK L+ ++ +GF P + +AQW HE ++ TS +
Sbjct: 226 LLCFQAPLDPNKMLLAGSS------LRGFSLPFLE--VAQWGLQHE*R*GFAINTSKIKK 71
Query: 146 GAAFG 150
G G
Sbjct: 70 GWLVG 56
>TC218649 similar to UP|Q39416 (Q39416) Integral membrane protein, partial
(93%)
Length = 1695
Score = 28.5 bits (62), Expect = 6.3
Identities = 28/131 (21%), Positives = 52/131 (39%), Gaps = 5/131 (3%)
Frame = +2
Query: 27 GFSIAYTVAADAA-----GINQSTKGTILSTFYYGYACSQVPGGYFAQKIGGRKILLFSF 81
GF+ YT +A G++ S S G + G A+ IG + L+ +
Sbjct: 218 GFTAGYTSPTQSAIINDLGLSVSEFSLFGSLSNVGAMVGAIASGQIAEYIGRKGSLMIAS 397
Query: 82 LLWSLTCALLPLDPNKTLILVTARLLVGVAQGFIFPSIHTVLAQWVPPHERSRSVSLTTS 141
+ + + + + + + RLL G G I ++ +A+ PP+ R VS+
Sbjct: 398 IPNIIGWLAISFAKDSSFLYM-GRLLEGFGVGIISYTVPVYIAEISPPNLRGGLVSVNQL 574
Query: 142 GMYLGAAFGML 152
+ +G L
Sbjct: 575 SVTIGIMLAYL 607
>TC214331 similar to UP|Q7NLY3 (Q7NLY3) Glr0986 protein, partial (14%)
Length = 1253
Score = 28.1 bits (61), Expect = 8.3
Identities = 15/44 (34%), Positives = 27/44 (61%), Gaps = 2/44 (4%)
Frame = +2
Query: 246 VVNNFTFHYALYVLMNWLPTYFELGLKLSLHEMGS--SKMLPYL 287
++N FH+ +YVL N LP++ + L ++ GS + +LP+L
Sbjct: 977 LINFQMFHHFIYVLFNLLPSHCFIILYIASPSSGSFCNLLLPFL 1108
>TC224181
Length = 597
Score = 28.1 bits (61), Expect = 8.3
Identities = 15/47 (31%), Positives = 23/47 (48%), Gaps = 1/47 (2%)
Frame = +2
Query: 126 WVPPHERSRSV-SLTTSGMYLGAAFGMLFLPSLVKFKGPQSVFVAES 171
W+ P+ +S S S+TTS M +L + F GP +F+ S
Sbjct: 47 WINPNAKSTSQDSVTTSAMLTNTMLLVLLRIFIRHFDGPDQIFIGNS 187
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.326 0.141 0.432
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 21,553,983
Number of Sequences: 63676
Number of extensions: 323106
Number of successful extensions: 2532
Number of sequences better than 10.0: 30
Number of HSP's better than 10.0 without gapping: 2503
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 2523
length of query: 443
length of database: 12,639,632
effective HSP length: 100
effective length of query: 343
effective length of database: 6,272,032
effective search space: 2151306976
effective search space used: 2151306976
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 15 ( 7.1 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.7 bits)
S2: 60 (27.7 bits)
Medicago: description of AC148651.2


