

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
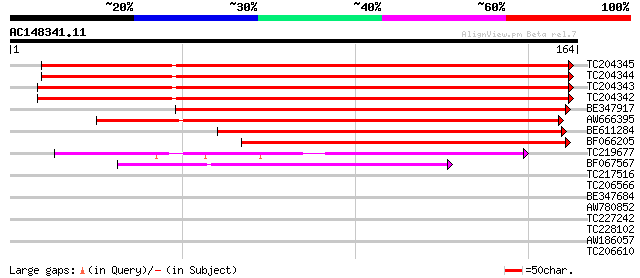
Query= AC148341.11 - phase: 0
(164 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC204345 homologue to UP|Q9XF99 (Q9XF99) Skp1 (Fragment), complete 256 4e-69
TC204344 homologue to UP|Q9XF99 (Q9XF99) Skp1 (Fragment), complete 252 5e-68
TC204343 homologue to UP|Q9XF99 (Q9XF99) Skp1 (Fragment), partia... 251 1e-67
TC204342 homologue to UP|Q9XF99 (Q9XF99) Skp1 (Fragment), partia... 248 7e-67
BE347917 similar to GP|4959710|gb| Skp1 {Medicago sativa}, parti... 165 1e-41
AW666395 similar to GP|18958257|db kinetochore protein {Brassica... 140 4e-34
BE611284 weakly similar to GP|4959710|gb|A Skp1 {Medicago sativa... 107 3e-24
BF066205 weakly similar to PIR|T10117|T101 fimbriata-associated ... 103 5e-23
TC219677 similar to GB|AAC28530.2|20197211|AC004665 E3 ubiquitin... 77 5e-15
BF067567 71 3e-13
TC217516 weakly similar to UP|Q8GUR8 (Q8GUR8) Thioredoxin h, par... 28 2.6
TC206566 weakly similar to UP|Q6VBI6 (Q6VBI6) Thioredoxin H2, pa... 27 3.4
BE347684 similar to GP|16519225|gb| cellulose synthase-like prot... 27 4.4
AW780852 weakly similar to GP|9965109|gb| resistance protein MG1... 27 5.7
TC227242 similar to UP|Q8GU90 (Q8GU90) PDR-like ABC transporter ... 27 5.7
TC228102 27 5.7
AW186057 weakly similar to GP|9280679|gb|A F2E2.11 {Arabidopsis ... 26 7.5
TC206610 weakly similar to UP|Q9XH49 (Q9XH49) Thioredoxin-like 4... 26 9.8
>TC204345 homologue to UP|Q9XF99 (Q9XF99) Skp1 (Fragment), complete
Length = 842
Score = 256 bits (653), Expect = 4e-69
Identities = 125/154 (81%), Positives = 140/154 (90%)
Frame = +2
Query: 10 ATKKITLKSSDNETFEIEKAVALESQTIKHLIDDNCADDSGIPLPNVTGKILAMVIEHCK 69
+TKKITLKSSD E FE+E+AVA+ESQTIKH+I+DNCAD SGIPLPNVT KILA VIE+CK
Sbjct: 128 STKKITLKSSDGEAFEVEEAVAVESQTIKHMIEDNCAD-SGIPLPNVTSKILAKVIEYCK 304
Query: 70 KHVDATSSDEKPSEDEINKWDTEFVKVDQDTLFDLILAANYLNIKSLLDLTCKTVADMIK 129
KHV+A +DEKPSEDE+ WD +FVKVDQ TLFDLILAANYLNIKSLLDLTC+TVADMIK
Sbjct: 305 KHVEANCADEKPSEDELKAWDADFVKVDQATLFDLILAANYLNIKSLLDLTCQTVADMIK 484
Query: 130 GRTPEEIRKTFNIINDYTPEEEEEVRSETQWAFD 163
G+TPEEIRKTFNI ND+TPEEEEEVR E QWAF+
Sbjct: 485 GKTPEEIRKTFNIKNDFTPEEEEEVRRENQWAFE 586
>TC204344 homologue to UP|Q9XF99 (Q9XF99) Skp1 (Fragment), complete
Length = 866
Score = 252 bits (644), Expect = 5e-68
Identities = 124/154 (80%), Positives = 139/154 (89%)
Frame = +2
Query: 10 ATKKITLKSSDNETFEIEKAVALESQTIKHLIDDNCADDSGIPLPNVTGKILAMVIEHCK 69
+TKKITLKSSD E FE+E+AVALESQTIKH+I+D+CAD SGIPLPNVT KILA VIE+CK
Sbjct: 104 STKKITLKSSDGEAFEVEEAVALESQTIKHMIEDDCAD-SGIPLPNVTSKILAKVIEYCK 280
Query: 70 KHVDATSSDEKPSEDEINKWDTEFVKVDQDTLFDLILAANYLNIKSLLDLTCKTVADMIK 129
KHV+A S+DEKPSED + WD +FV VDQ TLFDLILAANYLNIKSLLDLTC+TVADMIK
Sbjct: 281 KHVEANSADEKPSEDVLKAWDVDFVNVDQATLFDLILAANYLNIKSLLDLTCQTVADMIK 460
Query: 130 GRTPEEIRKTFNIINDYTPEEEEEVRSETQWAFD 163
G+TPEEIRKTFNI ND+TPEEEEEVR E QWAF+
Sbjct: 461 GKTPEEIRKTFNIKNDFTPEEEEEVRRENQWAFE 562
>TC204343 homologue to UP|Q9XF99 (Q9XF99) Skp1 (Fragment), partial (98%)
Length = 1090
Score = 251 bits (640), Expect = 1e-67
Identities = 122/155 (78%), Positives = 140/155 (89%)
Frame = +2
Query: 9 TATKKITLKSSDNETFEIEKAVALESQTIKHLIDDNCADDSGIPLPNVTGKILAMVIEHC 68
++ KKITLKSSD E FE+E+AVALESQTIKH+I+D+CAD SGIPLPNVT KILA VIE+C
Sbjct: 110 SSAKKITLKSSDGEAFEVEEAVALESQTIKHMIEDDCAD-SGIPLPNVTSKILAKVIEYC 286
Query: 69 KKHVDATSSDEKPSEDEINKWDTEFVKVDQDTLFDLILAANYLNIKSLLDLTCKTVADMI 128
KKHV+A + ++KPSED++ WD EFVKVDQ TLFDLILAANYLNIKSLLDLTC+TVADMI
Sbjct: 287 KKHVEAANPEDKPSEDDLKAWDAEFVKVDQATLFDLILAANYLNIKSLLDLTCQTVADMI 466
Query: 129 KGRTPEEIRKTFNIINDYTPEEEEEVRSETQWAFD 163
KG+TPEEIRKTFNI ND+TPEEEEEVR E QWAF+
Sbjct: 467 KGKTPEEIRKTFNIKNDFTPEEEEEVRRENQWAFE 571
>TC204342 homologue to UP|Q9XF99 (Q9XF99) Skp1 (Fragment), partial (98%)
Length = 852
Score = 248 bits (634), Expect = 7e-67
Identities = 120/155 (77%), Positives = 140/155 (89%)
Frame = +3
Query: 9 TATKKITLKSSDNETFEIEKAVALESQTIKHLIDDNCADDSGIPLPNVTGKILAMVIEHC 68
++ KKITLKSSD E FE+++AVALESQTIKH+I+D+CAD SGIPLPNVT KILA VIE+C
Sbjct: 78 SSAKKITLKSSDGEAFEVDEAVALESQTIKHMIEDDCAD-SGIPLPNVTSKILAKVIEYC 254
Query: 69 KKHVDATSSDEKPSEDEINKWDTEFVKVDQDTLFDLILAANYLNIKSLLDLTCKTVADMI 128
KKHV+A + ++KPSED++ WD +FVKVDQ TLFDLILAANYLNIKSLLDLTC+TVADMI
Sbjct: 255 KKHVEAANPEDKPSEDDLKAWDADFVKVDQATLFDLILAANYLNIKSLLDLTCQTVADMI 434
Query: 129 KGRTPEEIRKTFNIINDYTPEEEEEVRSETQWAFD 163
KG+TPEEIRKTFNI ND+TPEEEEEVR E QWAF+
Sbjct: 435 KGKTPEEIRKTFNIKNDFTPEEEEEVRRENQWAFE 539
>BE347917 similar to GP|4959710|gb| Skp1 {Medicago sativa}, partial (74%)
Length = 343
Score = 165 bits (417), Expect = 1e-41
Identities = 82/114 (71%), Positives = 94/114 (81%)
Frame = -1
Query: 49 SGIPLPNVTGKILAMVIEHCKKHVDATSSDEKPSEDEINKWDTEFVKVDQDTLFDLILAA 108
SGI P+VT KI VI+ CK HV+A + PSE ++ WD +FVKVDQDTLFDLILAA
Sbjct: 343 SGIHRPHVTSKIQQKVIK*CK*HVEAVDPEYNPSEYDL*AWDADFVKVDQDTLFDLILAA 164
Query: 109 NYLNIKSLLDLTCKTVADMIKGRTPEEIRKTFNIINDYTPEEEEEVRSETQWAF 162
+YLNIKSLLDLTC+TVADMIKG+TPEEIRKTF+I ND+TPEEEEEVR E QWAF
Sbjct: 163 HYLNIKSLLDLTCQTVADMIKGKTPEEIRKTFHIKNDFTPEEEEEVRRENQWAF 2
>AW666395 similar to GP|18958257|db kinetochore protein {Brassica juncea},
partial (63%)
Length = 436
Score = 140 bits (352), Expect = 4e-34
Identities = 76/135 (56%), Positives = 99/135 (73%)
Frame = +2
Query: 26 IEKAVALESQTIKHLIDDNCADDSGIPLPNVTGKILAMVIEHCKKHVDATSSDEKPSEDE 85
+ +A+ALES I HLI +CA +S IPLPNV ILA VI+ K+V A +++++PS DE
Sbjct: 35 VYEALALESHDIIHLIMLDCAGNS-IPLPNVASGILANVIKISWKYV*AENTEDEPS*DE 211
Query: 86 INKWDTEFVKVDQDTLFDLILAANYLNIKSLLDLTCKTVADMIKGRTPEEIRKTFNIIND 145
+ DT ++ +Q TL L L+ANYLNI +L+DL C TVADMIKG+TP+EIRKTFN ND
Sbjct: 212 LRC*DTNIMRGEQATLCHLFLSANYLNIDNLMDLACATVADMIKGKTPDEIRKTFNNKND 391
Query: 146 YTPEEEEEVRSETQW 160
+TPEEEEE R E +W
Sbjct: 392 FTPEEEEEGRRENKW 436
>BE611284 weakly similar to GP|4959710|gb|A Skp1 {Medicago sativa}, partial
(56%)
Length = 304
Score = 107 bits (267), Expect = 3e-24
Identities = 57/101 (56%), Positives = 71/101 (69%)
Frame = +2
Query: 61 LAMVIEHCKKHVDATSSDEKPSEDEINKWDTEFVKVDQDTLFDLILAANYLNIKSLLDLT 120
LA VI +CKK V+A +DEKPSE E+ WD +FV VDQ TLFDLIL A LN+ L DLT
Sbjct: 2 LAKVIYYCKKLVEAKCADEKPSEHELKAWDADFVNVDQATLFDLILVAIALNMLYLFDLT 181
Query: 121 CKTVADMIKGRTPEEIRKTFNIINDYTPEEEEEVRSETQWA 161
C+TVA MI GRT EI +TF I ND+T + +E+V + + A
Sbjct: 182 CQTVAYMIMGRTSVEIGETFYIQNDFTLDGDEDVLRDNRRA 304
>BF066205 weakly similar to PIR|T10117|T101 fimbriata-associated protein -
sweet orange (fragment), partial (76%)
Length = 286
Score = 103 bits (256), Expect = 5e-23
Identities = 52/95 (54%), Positives = 64/95 (66%)
Frame = -1
Query: 68 CKKHVDATSSDEKPSEDEINKWDTEFVKVDQDTLFDLILAANYLNIKSLLDLTCKTVADM 127
C++HV ++ E++ WD FV V Q T+FDLI+ + NIKS+LDLTC TV DM
Sbjct: 286 CQQHVRIVDPEDHFYEEDSKVWDDNFVTVAQATMFDLIMDXTHFNIKSVLDLTC*TVVDM 107
Query: 128 IKGRTPEEIRKTFNIINDYTPEEEEEVRSETQWAF 162
IKG+ EEIRKTFNI N + EEEEEV E QWAF
Sbjct: 106 IKGKILEEIRKTFNIKNAFILEEEEEVCWEXQWAF 2
>TC219677 similar to GB|AAC28530.2|20197211|AC004665 E3 ubiquitin ligase SCF
complex subunit SKP1/ASK1-related {Arabidopsis
thaliana;} , partial (62%)
Length = 478
Score = 76.6 bits (187), Expect = 5e-15
Identities = 51/146 (34%), Positives = 87/146 (58%), Gaps = 9/146 (6%)
Frame = +2
Query: 14 ITLKSSDNETFEIEKAVALESQTIKHLI-------DDNCADDSGIPLPN-VTGKILAMVI 65
+ L++SD+ ++E+ +A+ S I I NCA I LP V+ +L++++
Sbjct: 71 VWLQTSDDSIQQVEQEIAMFSPLICQEIIQKGMGSSKNCA----ICLPQQVSPAMLSLIL 238
Query: 66 EHCKKH-VDATSSDEKPSEDEINKWDTEFVKVDQDTLFDLILAANYLNIKSLLDLTCKTV 124
++C+ H V S+ E+ S DE +FV++D + L +L AA+ L +K L+DLT + +
Sbjct: 239 DYCRFHQVPGRSNKERKSYDE------KFVRIDTERLCELTSAADSLQLKPLVDLTSRAL 400
Query: 125 ADMIKGRTPEEIRKTFNIINDYTPEE 150
A +I+G+TPEEIR F++ +D T EE
Sbjct: 401 ARIIEGKTPEEIRDIFHLPDDLTEEE 478
>BF067567
Length = 289
Score = 70.9 bits (172), Expect = 3e-13
Identities = 44/97 (45%), Positives = 57/97 (58%)
Frame = +2
Query: 32 LESQTIKHLIDDNCADDSGIPLPNVTGKILAMVIEHCKKHVDATSSDEKPSEDEINKWDT 91
LES TI ++IDD+ K A I +C+KH A + + K +D IN DT
Sbjct: 2 LESHTINYMIDDHPPQQRHFSTEREE-KHPAYHI*YCEKHPLAPNPENKQCDDHINASDT 178
Query: 92 EFVKVDQDTLFDLILAANYLNIKSLLDLTCKTVADMI 128
EF KVD+ TLF LIL A+YL + +LLDL TVA+MI
Sbjct: 179 EFAKVDKGTLFYLILYAHYLGMNTLLDLISLTVANMI 289
>TC217516 weakly similar to UP|Q8GUR8 (Q8GUR8) Thioredoxin h, partial (58%)
Length = 764
Score = 27.7 bits (60), Expect = 2.6
Identities = 13/53 (24%), Positives = 28/53 (52%)
Frame = +2
Query: 87 NKWDTEFVKVDQDTLFDLILAANYLNIKSLLDLTCKTVADMIKGRTPEEIRKT 139
N D +F+K+D + L ++ A + + + + VAD + G EE++++
Sbjct: 314 NYTDVDFIKIDVEELTEVSQALQVYQLPTFILVKKGKVADRVVGVKKEELKRS 472
>TC206566 weakly similar to UP|Q6VBI6 (Q6VBI6) Thioredoxin H2, partial (78%)
Length = 921
Score = 27.3 bits (59), Expect = 3.4
Identities = 14/49 (28%), Positives = 25/49 (50%)
Frame = +1
Query: 90 DTEFVKVDQDTLFDLILAANYLNIKSLLDLTCKTVADMIKGRTPEEIRK 138
D EFVK+D D L ++ + + + + VAD + G EE+++
Sbjct: 331 DVEFVKIDVDELMEVSQHYQVQGMPTFMLIKKGNVADKVVGVRKEELQR 477
>BE347684 similar to GP|16519225|gb| cellulose synthase-like protein OsCslE1
{Oryza sativa}, partial (4%)
Length = 480
Score = 26.9 bits (58), Expect = 4.4
Identities = 16/71 (22%), Positives = 34/71 (47%), Gaps = 1/71 (1%)
Frame = +1
Query: 65 IEHCKKHVDATSSDEKPSEDEINKWDTEFVKVDQDTLFDLIL-AANYLNIKSLLDLTCKT 123
IE+ K + S+ + + ++WD+ + + D DT+ ++L N N K + T
Sbjct: 13 IEYAVK-LGGVPSESRSKHNGFSQWDSYYSRHDHDTILQILLHERNPHNSKDVDGFVLPT 189
Query: 124 VADMIKGRTPE 134
+ M + + P+
Sbjct: 190 LVYMAREKRPQ 222
>AW780852 weakly similar to GP|9965109|gb| resistance protein MG13 {Glycine
max}, partial (29%)
Length = 441
Score = 26.6 bits (57), Expect = 5.7
Identities = 13/39 (33%), Positives = 26/39 (66%), Gaps = 2/39 (5%)
Frame = -1
Query: 100 TLFDLILAANY--LNIKSLLDLTCKTVADMIKGRTPEEI 136
TL LI+ ++ L+ K +++ TC++VA ++ G+T + I
Sbjct: 174 TLQFLIIHEDFESLSAKGIIEKTCESVAGILSGKTQKNI 58
>TC227242 similar to UP|Q8GU90 (Q8GU90) PDR-like ABC transporter (PDR3 ABC
transporter), partial (17%)
Length = 1126
Score = 26.6 bits (57), Expect = 5.7
Identities = 11/18 (61%), Positives = 12/18 (66%)
Frame = -2
Query: 64 VIEHCKKHVDATSSDEKP 81
V EHC KH SS+EKP
Sbjct: 69 VPEHCSKHQGYESSEEKP 16
>TC228102
Length = 1679
Score = 26.6 bits (57), Expect = 5.7
Identities = 21/67 (31%), Positives = 29/67 (42%)
Frame = +1
Query: 34 SQTIKHLIDDNCADDSGIPLPNVTGKILAMVIEHCKKHVDATSSDEKPSEDEINKWDTEF 93
SQT +H D+ G+ P + L M EH + E PSE E+N E
Sbjct: 484 SQTSQHGHMDSSVSLEGLSTPRLDVNGL-MSSEHLNGAEGYANDKEHPSELELNHHSAED 660
Query: 94 VKVDQDT 100
VK++ T
Sbjct: 661 VKINTMT 681
>AW186057 weakly similar to GP|9280679|gb|A F2E2.11 {Arabidopsis thaliana},
partial (12%)
Length = 228
Score = 26.2 bits (56), Expect = 7.5
Identities = 12/30 (40%), Positives = 18/30 (60%)
Frame = -1
Query: 16 LKSSDNETFEIEKAVALESQTIKHLIDDNC 45
+K+S +E I+ + L S IKH+I D C
Sbjct: 228 VKTSADEEESIQLDLDLPSNNIKHVIQDTC 139
>TC206610 weakly similar to UP|Q9XH49 (Q9XH49) Thioredoxin-like 4, partial
(93%)
Length = 674
Score = 25.8 bits (55), Expect = 9.8
Identities = 15/49 (30%), Positives = 24/49 (48%)
Frame = +3
Query: 90 DTEFVKVDQDTLFDLILAANYLNIKSLLDLTCKTVADMIKGRTPEEIRK 138
D F+ +D D + ++ + + L L+ T D I G P+EIRK
Sbjct: 222 DVLFLTLDVDEVKEIASKMEIKAMPTFLLLSGGTPVDKIVGANPDEIRK 368
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.310 0.129 0.358
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 5,891,165
Number of Sequences: 63676
Number of extensions: 64122
Number of successful extensions: 265
Number of sequences better than 10.0: 36
Number of HSP's better than 10.0 without gapping: 259
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 259
length of query: 164
length of database: 12,639,632
effective HSP length: 90
effective length of query: 74
effective length of database: 6,908,792
effective search space: 511250608
effective search space used: 511250608
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.8 bits)
S2: 55 (25.8 bits)
Medicago: description of AC148341.11


