

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC147472.6 + phase: 0 /pseudo
(180 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
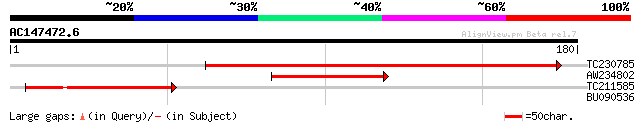
Score E
Sequences producing significant alignments: (bits) Value
TC230785 homologue to GB|AAO63344.1|28950841|BT005280 At1g02410 ... 218 1e-57
AW234802 71 3e-13
TC211585 similar to GB|AAO63344.1|28950841|BT005280 At1g02410 {A... 66 8e-12
BU090536 26 8.9
>TC230785 homologue to GB|AAO63344.1|28950841|BT005280 At1g02410 {Arabidopsis
thaliana;} , partial (41%)
Length = 857
Score = 218 bits (555), Expect = 1e-57
Identities = 103/113 (91%), Positives = 107/113 (94%)
Frame = +2
Query: 63 VVQFNADISDGMPWKFLPTQREVRVKPGESALAFYTVENQSSTPITGVSTYNVTPMKLGV 122
VVQFNADI+DGM WKF+PTQREVRVKPGESALAFYT EN+SSTPITGVSTYNVTPMK V
Sbjct: 2 VVQFNADIADGMQWKFVPTQREVRVKPGESALAFYTAENKSSTPITGVSTYNVTPMKAAV 181
Query: 123 YFNKIQCFCFEEQRLLPGEKIDMPVFFYIDPEIEDDPKMNGINNIILSYTFFK 175
YFNKIQCFCFEEQRLLPGE+IDMPVFFYIDPE E DPKMNGINNIILSYTFFK
Sbjct: 182 YFNKIQCFCFEEQRLLPGEQIDMPVFFYIDPEFETDPKMNGINNIILSYTFFK 340
>AW234802
Length = 419
Score = 70.9 bits (172), Expect = 3e-13
Identities = 33/37 (89%), Positives = 36/37 (97%)
Frame = +1
Query: 84 EVRVKPGESALAFYTVENQSSTPITGVSTYNVTPMKL 120
+VRVKPGESALAFYT EN+SSTPITGVSTYNVTPMK+
Sbjct: 10 QVRVKPGESALAFYTAENKSSTPITGVSTYNVTPMKV 120
>TC211585 similar to GB|AAO63344.1|28950841|BT005280 At1g02410 {Arabidopsis
thaliana;} , partial (16%)
Length = 144
Score = 66.2 bits (160), Expect = 8e-12
Identities = 34/48 (70%), Positives = 37/48 (76%)
Frame = +1
Query: 6 LTGLAFGMGVLTIFASPPLYRTFCQATSYDGTVTRRERVEEKIARHDS 53
LTGL F M T +A+ PLYR FCQAT Y GTVTRRE VEEKIARHD+
Sbjct: 4 LTGLVFAMVGCT-YAAVPLYRRFCQATGYGGTVTRRETVEEKIARHDT 144
>BU090536
Length = 418
Score = 26.2 bits (56), Expect = 8.9
Identities = 24/98 (24%), Positives = 35/98 (35%), Gaps = 23/98 (23%)
Frame = +2
Query: 96 FYTVENQSSTPITGVSTYNVTPMKL----------------------GVYFNKIQCFCFE 133
F T+ NQ I STY P L G Y NK +C C+
Sbjct: 119 FSTL*NQDEGSICSCSTYMPCPQPLLVRGVNGWFCGICCSKCNCVPSGTYGNKHECPCYR 298
Query: 134 EQRLLPGE-KIDMPVFFYIDPEIEDDPKMNGINNIILS 170
+ + G+ K FF+ P I P + + + +S
Sbjct: 299 DMKNSKGKAKCP*LFFFFHHPHINFKPLIQSLPCMYIS 412
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.322 0.138 0.411
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 7,571,634
Number of Sequences: 63676
Number of extensions: 89415
Number of successful extensions: 402
Number of sequences better than 10.0: 8
Number of HSP's better than 10.0 without gapping: 401
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 402
length of query: 180
length of database: 12,639,632
effective HSP length: 91
effective length of query: 89
effective length of database: 6,845,116
effective search space: 609215324
effective search space used: 609215324
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 56 (26.2 bits)
Medicago: description of AC147472.6


