

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
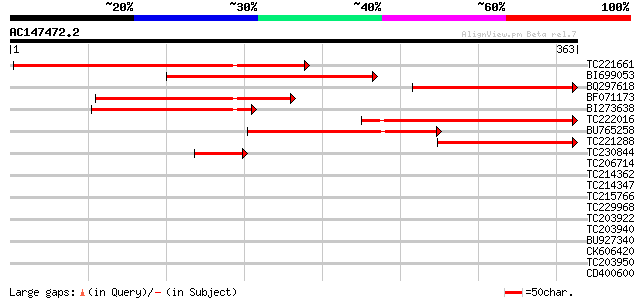
Query= AC147472.2 - phase: 0
(363 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC221661 weakly similar to UP|Q9SZ83 (Q9SZ83) AX110P-like protei... 240 6e-64
BI699053 212 2e-55
BQ297618 184 4e-47
BF071173 weakly similar to PIR|T14319|T14 protein AX110P - carro... 147 6e-36
BI273638 144 6e-35
TC222016 weakly similar to UP|Q9SZ83 (Q9SZ83) AX110P-like protei... 124 7e-29
BU765258 weakly similar to PIR|T14319|T14 protein AX110P - carro... 120 7e-28
TC221288 weakly similar to UP|Q9SZ83 (Q9SZ83) AX110P-like protei... 84 8e-17
TC230844 42 3e-04
TC206714 similar to UP|Q6H436 (Q6H436) Oxidoreductase family pro... 38 0.006
TC214362 weakly similar to UP|Q7K0Y2 (Q7K0Y2) LD23217p, partial ... 33 0.20
TC214347 homologue to UP|Q43821 (Q43821) Ubiquitin conjugating e... 33 0.20
TC215766 similar to UP|Q93XA6 (Q93XA6) NAC domain protein NAC2, ... 32 0.45
TC229968 similar to UP|O64797 (O64797) T1F15.5 protein, partial ... 30 1.3
TC203922 homologue to UP|Q7X9L1 (Q7X9L1) Holocarboxylase synthet... 29 2.9
TC203940 29 3.8
BU927340 similar to GP|18087570|gb AT3g54650/T5N23_10 {Arabidops... 28 5.0
CK606420 28 6.5
TC203950 28 8.5
CD400600 homologue to PIR|A84710|A847 photosystem II reaction ce... 28 8.5
>TC221661 weakly similar to UP|Q9SZ83 (Q9SZ83) AX110P-like protein, partial
(51%)
Length = 567
Score = 240 bits (613), Expect = 6e-64
Identities = 114/190 (60%), Positives = 146/190 (76%)
Frame = +3
Query: 3 EKDTVRFGILGCATIARKLARAITLTPNATISAIGSRSISKAAKFAVENSLPASVRIYGS 62
E T+RFG++GCA IARK++RAI+L PNA + A+GSRS+ KA FA N PA+ +++GS
Sbjct: 3 ETPTIRFGVIGCADIARKVSRAISLAPNAALYAVGSRSLDKARAFAAANGFPAAAKVHGS 182
Query: 63 YDEVVEDSGVDAVYIPLPTSLHVRWAVAAANKKKHVLVEKPAALDVAELDLILEACEANG 122
Y+ V++D VDAVY+PLPTSLHVRWAV AA KKKHVL+EKP AL AE D I+EACE+NG
Sbjct: 183 YEAVLDDPDVDAVYVPLPTSLHVRWAVLAAQKKKHVLLEKPVALHAAEFDEIVEACESNG 362
Query: 123 VQFMDGCMWLHHPRTSNMKHLLDLSNSDGIGPVQFIHSTSTMPTTQEFLENNIRVKPELD 182
VQ MDG MW+HHPRT+ M+ LS+S+ G ++ IH+ T +FLEN+IRVKP+LD
Sbjct: 363 VQLMDGTMWMHHPRTAKMRDF--LSDSNRFGQLRSIHTHFTFAADADFLENDIRVKPDLD 536
Query: 183 ALGALGDLAW 192
ALG+LGD W
Sbjct: 537 ALGSLGDEGW 566
>BI699053
Length = 408
Score = 212 bits (540), Expect = 2e-55
Identities = 102/136 (75%), Positives = 119/136 (87%), Gaps = 1/136 (0%)
Frame = +1
Query: 101 EKPAALDVAELDLILEACEANGVQFMDGCMWLHHPRTSNMKHLLDLSNS-DGIGPVQFIH 159
EKP ALDVAELD IL+A E+NGVQFMDG MWLHHPRT++++ L + +S + +GPV+FIH
Sbjct: 1 EKPVALDVAELDRILDAVESNGVQFMDGSMWLHHPRTAHIQRLFSVPHSANSVGPVRFIH 180
Query: 160 STSTMPTTQEFLENNIRVKPELDALGALGDLAWYCIGASLWAKGYKLPTTVTALPDVTRN 219
STSTMP T EFLE++IRVKPELD LGALGDLAWYCIGASLWAKGY+LPT+VTALPD TRN
Sbjct: 181 STSTMPATPEFLESDIRVKPELDGLGALGDLAWYCIGASLWAKGYQLPTSVTALPDSTRN 360
Query: 220 MAGVILSITTSLQWDQ 235
+GVI+SIT SL WDQ
Sbjct: 361 DSGVIVSITASLLWDQ 408
>BQ297618
Length = 420
Score = 184 bits (468), Expect = 4e-47
Identities = 90/105 (85%), Positives = 95/105 (89%)
Frame = +1
Query: 259 GSNGSMHLKDFIIPYKETSASFDFTFGAKFVDLHIGWNVRPEEVHVVNKLPQEALMVQEF 318
GSNGS+HL+DFIIPY ETSASFD T GAKFVDLHIGWNVRPEEVHV N+LPQEALMVQEF
Sbjct: 1 GSNGSLHLRDFIIPYGETSASFDLTLGAKFVDLHIGWNVRPEEVHVANELPQEALMVQEF 180
Query: 319 AMLVASIRDSGSQPSIKWPEISRKTQLVVDAVKKSLELGCKPVSL 363
+ LVA IRD GS+PS KWPEISRKTQLVVDAV KSLELGCKPV L
Sbjct: 181 SRLVAGIRDCGSKPSTKWPEISRKTQLVVDAVIKSLELGCKPVCL 315
>BF071173 weakly similar to PIR|T14319|T14 protein AX110P - carrot, partial
(32%)
Length = 381
Score = 147 bits (372), Expect = 6e-36
Identities = 69/128 (53%), Positives = 94/128 (72%)
Frame = +3
Query: 56 SVRIYGSYDEVVEDSGVDAVYIPLPTSLHVRWAVAAANKKKHVLVEKPAALDVAELDLIL 115
S ++YGSY+ ++ED VDAVY+PLPTSLH+ WAV AA KKH+L+EKP ALDVA+ D IL
Sbjct: 3 SAKVYGSYESLLEDPDVDAVYMPLPTSLHLHWAVLAAQNKKHLLLEKPVALDVAQFDRIL 182
Query: 116 EACEANGVQFMDGCMWLHHPRTSNMKHLLDLSNSDGIGPVQFIHSTSTMPTTQEFLENNI 175
EACE++GVQFMD MW+H+PRT+ M H L+++ G ++ + + T ++L NI
Sbjct: 183 EACESSGVQFMDNTMWVHNPRTAAMAHF--LNDAQRFGNLKSVRTCFTFAADPDYLNTNI 356
Query: 176 RVKPELDA 183
RVK +LDA
Sbjct: 357 RVKSDLDA 380
>BI273638
Length = 358
Score = 144 bits (363), Expect = 6e-35
Identities = 68/106 (64%), Positives = 85/106 (80%)
Frame = +2
Query: 53 LPASVRIYGSYDEVVEDSGVDAVYIPLPTSLHVRWAVAAANKKKHVLVEKPAALDVAELD 112
LP +VR+YGSY+ V+ED VDAVYIPLPT LHV WAV AA ++KHVL+EKP A++VAELD
Sbjct: 5 LPEAVRVYGSYEGVLEDEEVDAVYIPLPTGLHVTWAVRAAERRKHVLLEKPVAMNVAELD 184
Query: 113 LILEACEANGVQFMDGCMWLHHPRTSNMKHLLDLSNSDGIGPVQFI 158
ILEACEA GVQFMDG MW+HHPRT+ MK LS++ G ++++
Sbjct: 185 RILEACEARGVQFMDGTMWVHHPRTAKMKEA--LSDAQRFGQLKWV 316
>TC222016 weakly similar to UP|Q9SZ83 (Q9SZ83) AX110P-like protein, partial
(35%)
Length = 656
Score = 124 bits (311), Expect = 7e-29
Identities = 66/138 (47%), Positives = 88/138 (62%)
Frame = +2
Query: 226 SITTSLQWDQPNQTVANIHCSFHSHTSMDLSISGSNGSMHLKDFIIPYKETSASFDFTFG 285
S SL W+ VA HCSF ++ +MD++ G+ G++H+ DFIIPY+E ASF
Sbjct: 2 SCGASLHWEDGK--VATFHCSFLTNLTMDITAVGTKGTLHVHDFIIPYEEKEASFYAATE 175
Query: 286 AKFVDLHIGWNVRPEEVHVVNKLPQEALMVQEFAMLVASIRDSGSQPSIKWPEISRKTQL 345
F DL W +P + + +PQEALMV EFA LVA I+ + S+P KWP ISRKTQL
Sbjct: 176 TGFDDLVSKWIPQPSKHIIKTDIPQEALMVTEFARLVADIKFNNSKPEKKWPTISRKTQL 355
Query: 346 VVDAVKKSLELGCKPVSL 363
V+DAVK S+E G +PV +
Sbjct: 356 VLDAVKASIERGFEPVQI 409
>BU765258 weakly similar to PIR|T14319|T14 protein AX110P - carrot, partial
(32%)
Length = 423
Score = 120 bits (302), Expect = 7e-28
Identities = 57/124 (45%), Positives = 80/124 (63%)
Frame = +3
Query: 153 GPVQFIHSTSTMPTTQEFLENNIRVKPELDALGALGDLAWYCIGASLWAKGYKLPTTVTA 212
G +++IHS T EFL+N+IRVKP+LD LGALGD WYCI A LWA Y+LP +V A
Sbjct: 18 GQLKWIHSCLTYNPGPEFLKNSIRVKPDLDGLGALGDTGWYCIRAILWAVNYELPKSVLA 197
Query: 213 LPDVTRNMAGVILSITTSLQWDQPNQTVANIHCSFHSHTSMDLSISGSNGSMHLKDFIIP 272
P N GVI+S +S+ W+ A HCSF S+ + D+++ G+ G + L DF +P
Sbjct: 198 FPGAILNEEGVIISCGSSMHWEDGRS--ATFHCSFLSYVTFDVTVLGTKGCLRLNDFALP 371
Query: 273 YKET 276
++E+
Sbjct: 372 FEES 383
>TC221288 weakly similar to UP|Q9SZ83 (Q9SZ83) AX110P-like protein, partial
(22%)
Length = 600
Score = 84.3 bits (207), Expect = 8e-17
Identities = 44/89 (49%), Positives = 58/89 (64%)
Frame = +1
Query: 275 ETSASFDFTFGAKFVDLHIGWNVRPEEVHVVNKLPQEALMVQEFAMLVASIRDSGSQPSI 334
E ASF F DL GW+ +P + V LPQEAL+++EFA LVA I+ S+P
Sbjct: 4 EKEASFLAGTETGFNDLVTGWDKQPSKHTVTTDLPQEALLIREFARLVAEIKFKNSKPDK 183
Query: 335 KWPEISRKTQLVVDAVKKSLELGCKPVSL 363
KWP I+RKTQLV+DAVK S++ G +PV +
Sbjct: 184 KWPTITRKTQLVLDAVKASIQRGFEPVQI 270
>TC230844
Length = 617
Score = 42.4 bits (98), Expect = 3e-04
Identities = 17/34 (50%), Positives = 25/34 (73%)
Frame = +2
Query: 119 EANGVQFMDGCMWLHHPRTSNMKHLLDLSNSDGI 152
E++GVQFMD +W+H+PRT+ M H L+ + GI
Sbjct: 374 ESSGVQFMDSTIWVHNPRTATMAHFLNDAQRFGI 475
>TC206714 similar to UP|Q6H436 (Q6H436) Oxidoreductase family protein-like,
partial (66%)
Length = 1401
Score = 38.1 bits (87), Expect = 0.006
Identities = 34/124 (27%), Positives = 56/124 (44%), Gaps = 7/124 (5%)
Frame = +2
Query: 1 MTEKDTVRFGILGCATIARK--LARAITLTPNATISAIGSRSISKAAKFAVE---NSLPA 55
M K + ILG + L R ++ + +I SR+ ++A AV+ P
Sbjct: 74 MATKQLPQIAILGAGIFVKTQYLPRLSEISHLFILKSIWSRT-QESATAAVDIARKHFPQ 250
Query: 56 SVRIYGS--YDEVVEDSGVDAVYIPLPTSLHVRWAVAAANKKKHVLVEKPAALDVAELDL 113
+G +D+++ D +DAV + L V ++ KHVL EKPAA +EL+
Sbjct: 251 VETKWGDNGFDDIIHDGSIDAVAVVLAGQNQVDISLRLLKAGKHVLQEKPAASCTSELEA 430
Query: 114 ILEA 117
L +
Sbjct: 431 ALSS 442
>TC214362 weakly similar to UP|Q7K0Y2 (Q7K0Y2) LD23217p, partial (4%)
Length = 685
Score = 33.1 bits (74), Expect = 0.20
Identities = 21/56 (37%), Positives = 26/56 (45%), Gaps = 3/56 (5%)
Frame = +3
Query: 164 MPTTQEFLENNIRVKPELDALGALGDLAWY---CIGASLWAKGYKLPTTVTALPDV 216
MPT +EFLE + VK LD G LG + Y + W K+ LPDV
Sbjct: 354 MPTWKEFLERIVEVKRLLDLAGLLGSIESYHGMLVSDVGWISWRKILGLALLLPDV 521
>TC214347 homologue to UP|Q43821 (Q43821) Ubiquitin conjugating enzyme ,
partial (36%)
Length = 899
Score = 33.1 bits (74), Expect = 0.20
Identities = 20/55 (36%), Positives = 27/55 (48%), Gaps = 2/55 (3%)
Frame = +3
Query: 164 MPTTQEFLENNIRVKPELDALGALGDLAWY--CIGASLWAKGYKLPTTVTALPDV 216
MPT +EFLE + VK LD G LG ++++ + W K LPDV
Sbjct: 618 MPTWKEFLERIVEVKRLLDLAGLLGRISYHGMLVSGVGWLSWRKTLGLALLLPDV 782
>TC215766 similar to UP|Q93XA6 (Q93XA6) NAC domain protein NAC2, partial
(89%)
Length = 1237
Score = 32.0 bits (71), Expect = 0.45
Identities = 24/87 (27%), Positives = 40/87 (45%)
Frame = +2
Query: 87 WAVAAANKKKHVLVEKPAALDVAELDLILEACEANGVQFMDGCMWLHHPRTSNMKHLLDL 146
W + KKK++ A + + L + Q M + PRTS++ HLL++
Sbjct: 614 WVLCRIYKKKNIGKSMEAKEEYPIAQINLTPANNDSEQEM-----VKFPRTSSLTHLLEM 778
Query: 147 SNSDGIGPVQFIHSTSTMPTTQEFLEN 173
D +GP+ I S +T +T +F N
Sbjct: 779 ---DYLGPISHILSDATYNSTFDFQIN 850
>TC229968 similar to UP|O64797 (O64797) T1F15.5 protein, partial (13%)
Length = 902
Score = 30.4 bits (67), Expect = 1.3
Identities = 19/54 (35%), Positives = 28/54 (51%), Gaps = 5/54 (9%)
Frame = +3
Query: 236 PNQTVANIHCSFHSHTSMDLSISGSNGSMH-----LKDFIIPYKETSASFDFTF 284
PN T+ NI C F + +S S S S+ S+H L +F +++SF TF
Sbjct: 21 PNPTIINILCFFQTPSSSSSSSSSSSSSIHSLLHTLHNFSSSESASASSFY*TF 182
>TC203922 homologue to UP|Q7X9L1 (Q7X9L1) Holocarboxylase synthetase
(Fragment), partial (7%)
Length = 560
Score = 29.3 bits (64), Expect = 2.9
Identities = 27/95 (28%), Positives = 41/95 (42%), Gaps = 14/95 (14%)
Frame = +2
Query: 264 MHLKDF----IIPYKETSASFDF-----TFGAKFVDLHIG-----WNVRPEEVHVVNKLP 309
M LKD I+P+ T +SFDF TF K+ + W P++ H+V
Sbjct: 236 MDLKDDAVKGIVPWDGTLSSFDFSLSAHTFSDKWKRFNPSFPAWKWITCPKQHHLVASHK 415
Query: 310 QEALMVQEFAMLVASIRDSGSQPSIKWPEISRKTQ 344
E + E + S ++ S S+ W E S +Q
Sbjct: 416 VEGYLSLENMCHIKSSQEQESDRSLTWKEESNISQ 520
>TC203940
Length = 932
Score = 28.9 bits (63), Expect = 3.8
Identities = 21/73 (28%), Positives = 36/73 (48%), Gaps = 2/73 (2%)
Frame = +3
Query: 61 GSYDEVVEDSGVDAVYIPLPTS--LHVRWAVAAANKKKHVLVEKPAALDVAELDLILEAC 118
GS+ VV DS V +V+ L + + + W +AA + V++ +LD++ E C
Sbjct: 468 GSFGFVVYDSKVGSVFAALGSDGGIKLYWGIAA---DESVVISD-------DLDVMKEGC 617
Query: 119 EANGVQFMDGCMW 131
+ F GCM+
Sbjct: 618 AKSFAPFPPGCMF 656
>BU927340 similar to GP|18087570|gb AT3g54650/T5N23_10 {Arabidopsis
thaliana}, partial (23%)
Length = 446
Score = 28.5 bits (62), Expect = 5.0
Identities = 13/30 (43%), Positives = 19/30 (63%)
Frame = -3
Query: 28 TPNATISAIGSRSISKAAKFAVENSLPASV 57
TP ++A+ ISK AKF EN++ AS+
Sbjct: 108 TPLIRLTAVSETEISKNAKFRHENAIQASI 19
>CK606420
Length = 498
Score = 28.1 bits (61), Expect = 6.5
Identities = 9/30 (30%), Positives = 16/30 (53%)
Frame = -3
Query: 179 PELDALGALGDLAWYCIGASLWAKGYKLPT 208
P++ G L W+C+G+ W G+ P+
Sbjct: 145 PKVSGSSPGGGLGWFCLGSFFWGGGFGDPS 56
>TC203950
Length = 1217
Score = 27.7 bits (60), Expect = 8.5
Identities = 22/73 (30%), Positives = 35/73 (47%), Gaps = 2/73 (2%)
Frame = +2
Query: 61 GSYDEVVEDSGVDAVYIPLPTSLHVR--WAVAAANKKKHVLVEKPAALDVAELDLILEAC 118
GS+ VV DS V +V+ L + V+ W +AA V++ +L++I E C
Sbjct: 458 GSFAFVVYDSKVGSVFAALGSDGGVKLYWGIAADGS---VVISD-------DLEVIKEGC 607
Query: 119 EANGVQFMDGCMW 131
+ F GCM+
Sbjct: 608 AKSFAPFPTGCMF 646
>CD400600 homologue to PIR|A84710|A847 photosystem II reaction center 6.1KD
protein [imported] - Arabidopsis thaliana, partial (42%)
Length = 553
Score = 27.7 bits (60), Expect = 8.5
Identities = 31/120 (25%), Positives = 47/120 (38%), Gaps = 9/120 (7%)
Frame = -3
Query: 72 VDAVYIPLPTSLHVRWAVAAANKKKHVLVEKPAALDVAELDLILEACEANGVQFMDGCMW 131
V+ + P PT ++ + KKK + +K D L L L A G Q +W
Sbjct: 458 VNYLVYPNPT*CYLLFTKKKKKKKKKIKKKKKKTRD*FSLSLSLVPNSARGCQAQQPWLW 279
Query: 132 LHHPRTSNMKHL---LDLSNSDGIGPVQF------IHSTSTMPTTQEFLENNIRVKPELD 182
L R S +K L L L+ + +G + S ST+P + + KP D
Sbjct: 278 L--MRDSALKELGFPLGLATTFLVGSCLVSLVSYGLSSXSTLPPLKRMKSLDCPSKPPCD 105
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.318 0.132 0.393
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 15,685,896
Number of Sequences: 63676
Number of extensions: 204113
Number of successful extensions: 1062
Number of sequences better than 10.0: 41
Number of HSP's better than 10.0 without gapping: 1055
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1058
length of query: 363
length of database: 12,639,632
effective HSP length: 98
effective length of query: 265
effective length of database: 6,399,384
effective search space: 1695836760
effective search space used: 1695836760
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 60 (27.7 bits)
Medicago: description of AC147472.2


